Research Article
Evaluation of miRNA-135a and miRNA-9 as Prognostic Markers in Breast Cancer Patients
1Department of Clinical Pathology, National Institute for Diabetes and Endocrinology, Cairo, Egypt
2Lecturer of Surgical Oncology, Faculty of Medicine, Suez Canal University, Egypt
3Department of Biochemistry, Faculty of Science, Suez Canal University, Egypt
4Lecturer of Surgical Oncology, Faculty of Medicine, Suez Canal University, Egypt
5Cardiothoracic Surgery, Faculty of Medicine, Al Azahar University, Egypt
6Department of Biochemistry, Faculty of Science, Fayoum University, Egypt
7Depatment of Zoology, Girls Collage for Arts, Science and Education, Ain Shams University, Egypt
8Kasr Al-Ainy Center of Clinical Oncology and Nuclear Medicine (NEMROCK), Kasr Al-Ainy School of Medicine, Cairo University, Egypt
9Clinical Oncology and Palliative Care Specialist, ARAR Oncology Center, Prince Abdulaziz bin Musaed Hospital, ARAR, Kingdom of Saudi Arabia
*Corresponding author: Mohamed M Gamea, Clinical Oncology and Palliative Care Specialist, ARAR Oncology Center, Prince Abdulaziz bin Musaed Hospital, ARAR, Kingdom of Saudi Arabia, E-mail: mohammed.game3@hotmail.com
Received: February 11, 2019 Accepted: March 1, 2019 Published: March 11, 2019
Citation: Hussein G, Faisal M, Kamal M, et al. Evaluation of miRNA-135a and miRNA-9 as Prognostic Markers in Breast Cancer Patients. Madridge J Oncogenesis. 2019; 3(1): 59-64. doi: 10.18689/mjo-1000111
Copyright: © 2019 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Background: Breast cancer remains one of the top threats to women health. The current lack of tumor markers with desirable sensitivity and specificity arises to be a major obstacle towards future management of breast cancer. Many studies are directed to reveal the diagnostic and prognostic potentials of circulating miRNAs in breast cancer. Aim: We attempt to evaluate the feasibility and clinical utility of circulating miRNA-135 and miRNA-9as prognostic biomarkers for breast cancer.
Methods: Methods was used Levels of miRNA-135 and miRNA-9 in sera from 126 participants representing two different groups, 40 healthy group and 86 breast cancer patients.
Results: Expression of both miR-135 and miR-9 were evaluated in both groups. Regarding miR-9, its expression was higher in case than in control group, however there were no significant differences in expression between the case and control groups when compared regarding age, menopausal status and BMI. Expression of miR-135 was higher in case group than control group, by comparing the two groups regarding age, menopausal status and BMI, there were significant differences. Higher miR-135 expression was associated with older age (age>35 years), post-menopausal and obese women in case group.
The miR-135levels of 27.59 or lesser provided a sensitivity of 90.70%, a specificity of 90% and a positive predictive value of 95.12%, negative predictive value of 81.82 %. According to these results, miR-135 had a good predictive ability. The miR-9 levels of 27.59 or lesser provided a sensitivity of 75.58%, a specificity of 80.00% and a positive predictive value of 89.04%, negative predictive value of 60.38%.
Conclusions: Our study establishes the role of miRNA 135 and miRNA 9 in breast cancer patientʼs disease progression and prognosis.
Keywords: Breast Cancer, miRNA-135, miRNA-9.
Introduction
Globally, breast cancer is the second most common cancer and it is the most frequent type of cancers among females [1]. It was reported that that the incidence of breast cancer in 2017 among female was 252,710 cases worldwide, with number of deaths reached 40,610 cases [2].The incidence rate in Egypt was found to be 29.9/100,000 population in the age group of 30-34 years [3].
The etiology of breast cancer involves the interaction between inherited genetic factors and environmental factors with a relative importance ranging from strong environmental or strong genetic factors [4]. Carcinogenesis occurs as multistep process which involves accumulation of genetic and epigenetic variations in the cell, this in turn leads to the progressive transformation of normal cell to cancer cell through initiation, promotion and progression steps [4].
The primary management of breast cancer is surgery; however this depends on the biological characteristics and stage of the tumor. Surgery involves several types such as mastectomy, modified radical mastectomy and lumpectomy with axillary lymph node dissection. Surgery can either be performed before or after receiving systemic therapy. The two principles of treatment aim to reduce the risk of local recurrence and the chance of metastasis. Early breast cancer is treated by surgery with or without involving of radiotherapy, if there are adverse prognostic factors such as lymph node involvement; systemic therapy (chemotherapy or hormone therapy) is added as there is an indication of high possibility of metastatic relapse [5].
Micro RNAs (miRNAs) are non-coding small cellular RNAs which composed of 17-27 base pair; they regulate gene expression post transcriptionally by the induction of translational repression or degradation of target mRNAs [6]. Expression of miRNAs has been associated with several human diseases including cancers as they have oncogenic properties where they targeting oncogenes [6]. miRNA can repress or promote cancer initiation and progression, the deregulation of miRNA expression is a common characteristic in these process [7-9].
Abnormal expression of miRNAs has been studied in different cancers including breast cancer [10,11]. It was reported that miRNA-135a was involved in hypertension, colorectal cancer, bone and muscle development [12-14] epithelial ovarian cancer [15] portal vein tumor thrombus and gastric cancer [16,17], however there is limitation in knowledge about its role in breast cancer.
One study showed that miR-135a level was elevated in breast cancer in case of metastasis, where it was found that miR-135a promoted the migration and invasiveness of breast cancer cells in vitro [6]. Another study showed that expression of miR-135a was decreased strongly in metastatic breast cancers when compared to non-metastatic ones [18].
MiR-9 targets directly CDH1 in breast cancer cells leading to increase in the invasiveness and cell motility [19]. It was reported that miR-9 was associated significantly with breast cancer local recurrence [20]. Another report showed that miR9 was elevated significantly in primary breast cancers patients who diagnosed with metastasis than in patients without metastasis [19].
Patients and Methods
Ethics statement
This research has been reviewed by our Research ethics committee in Faculty of medicine-Suez Canal University at its meeting on 23/2/2016 with reference number (#4162). A written and verbal informed consent was taken from the selected patients.
Study design and sample collection
In the present study, this cross-section study included 40 age-matched healthy female as a control group while the case group included 86 female patients were collected from surgical oncology department of Suez Canal University in Ismailia city, Egypt from march 2016 to January 2018.
Participants of both groups for this study didnʼt suffer any chronic disease, in the case group there were 9 cases who had breast cancer and whose age was less than 35 years old. Immunohistochemistry was used to assess hormone receptors status, for ER and PR receptors, the positive cases were identified when atleast 1% of the invasive carcinoma cells displayed nuclear reactivity, while for HER2 the positive cases were identified when they scored 3+ or scored 2+ with confirmation by validated in situ hybridization technique.
Micro RNA extraction
MiRNA was isolated from sera by using the miRNeasy Mini kit (QIAgen, Germany) according to manufacturerʼs instructions. Then finally, miRNA was recovered in 30 uL of RNase-free water.
Statistical methods
Statistical analysis was performed with the statistical software SPSS 23.0 (SPSS Inc., Chicago, IL, USA). Quantitative variables that followed a normal distribution were defined by the mean and standard deviation. For non-Gaussian variables, the median and range were used. Qualitative variables were defined by number and percentage of cases. Comparison of quantitative variables was performed with a Studentʼs t-test meanwhile for qualitative variables the Chi-square test were performed. In those cases with fewer than 5 observations in the cell, the Fisher exact probability method was used. P<0.05 was regarded as significant.
Results
The present study included 40 healthy females as control group, while the case group included 86 female patients.
In the case group, there were 56(65.1%) of females with pre-menopausal, while 30(34.9%) were post-menopausal.
In case group had a positive family history of breast cancer 19(22.1%) and 24(27.9%) were suffering co-morbidity and the same percent had a multicentric tumor.
Of 86 cases, there were 34(39.5%) performed conservative surgery, 44(51.2%) performed modified radical surgery and only 8(9.3%) didnʼt perform any surgery.
According to pathology examination, the large majority 69(80.2%) had invasive ductal carcinoma (IDC), whereas 10(11.6%) had invasive lobular carcinoma (ILC) and 7(8.1%) only suffered both. The most common grade among our patients was grade 2 which was found in 48(55.8%) followed by grade 3 which represented 26(30.2%) among patients and grade 1 was found in 12(14%).
Progesterone receptor (PR) was present in 43(50%) of cases, while Estrogen receptor (ER) was present in 42(48.8%) and Human epidermal growth factor receptor 2 (HER2) was present in 23(26.7%). The molecular sub-typing of breast cancer of patients showed that luminal B was the most common among patients representing 36(41.9%), followed by those with triple negative 16(18.6%) then luminal A and HER representing 15(17.4%) and 12(14%) respectively, however 7(8.1%) couldnʼt be determined.
Most of our patients had breast cancer of III stage 36(41.9%) followed by those with stage II 31(36%) and finally those with stage I and IV representing 11(12.8%) and 8(9.3%) respectively.
There were 10 cases suffered metastasis; most of them 7(8.1%) suffered liver metastasis, 2(2.3%) suffered lung metastasis and 1(1.2%) suffered bone metastasis. Tumor size of T2 was 51(59.3%); 21 (24.4%) were T1 and 14(16.3%) were T3.
Lymph node involvement (N0) 28 (32.6%, 22(25.6%) had one lymph node involvement (N1), 30(34.9%) and 6(7%) had 2 (N2) and 3(N3) lymph node involvement.
Regarding age of all participants, the mean ± SD age of control group was 37.98 ± 10.45, while for case group, it was 46.81 ± 87, and there was a significant difference between the two groups (P-value<0.01).
A comparison between the two groups was made regarding age, menopausal status and BMI shown in table 1. There were significant differences regarding the three variables between the two groups, the case group tended to be older in age and higher in BMI than control; however both groups tended to be in the pre-menopausal time.
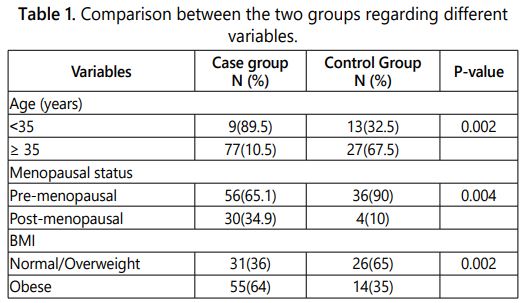
Expression of both miR-135 and miR-9 were evaluated in both groups. Regarding miR-9, its expression was higher in case than in control group, however there were no significant differences in expression between the case and control groups when compared regarding age, menopausal status and BMI (Table 2).
Expression of miR-135 was higher in case group than control group, by comparing the two groups regarding age, menopausal status and BMI, there were significant differences. Higher miR-135 expression was associated with older age (age>35 years), post-menopausal and obese women in case group (Table 2).
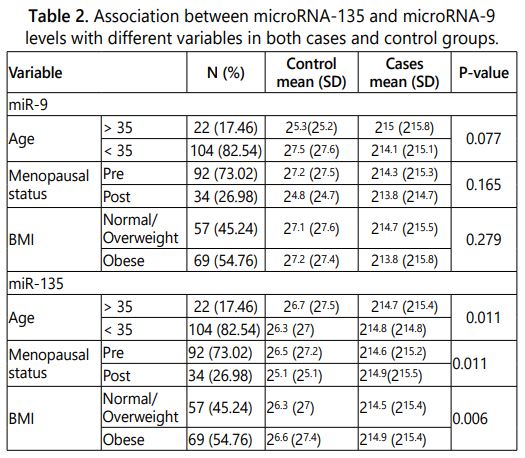
In case group, the effect of several variables on the expression of miR-9 and miR-135 were investigated. Age, BMI, metastasis, ER, PR and Her2n receptors as well as pathological type of tumor didnʼt affect the expression of the studies microRNAs.
The tumor size and stage significantly affected the expression of the two microRNAs under study, tumor size 3 was significantly associated with over expression of both miR-135 (215.5) (P-value<0.01) and miR-9 (215.3) (P-value=0.004). Stage III of the tumor was significantly associated with over expression of both miR-135 (215) (P-value=0.001) and miR-9 (215.2) (P-value<0.01).
The tumor grade significantly (P-value<0.01) affected the level of miR-135, grade 3 was associated with the over expression (216).
The lymph node involvement was significantly (P-value<0.01) affected the expression of miR-9 only, where grade 3 involvements were associated with miR-9 over expression (216.4).
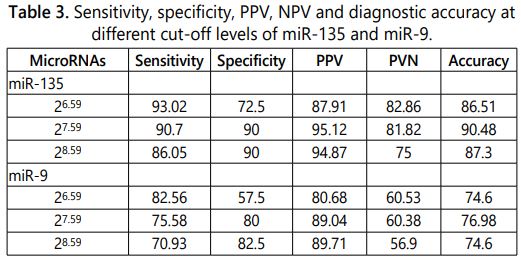
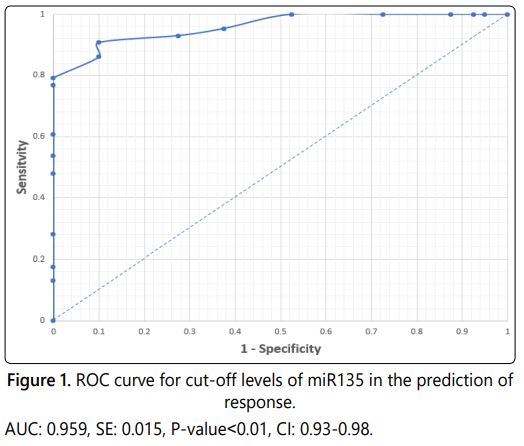
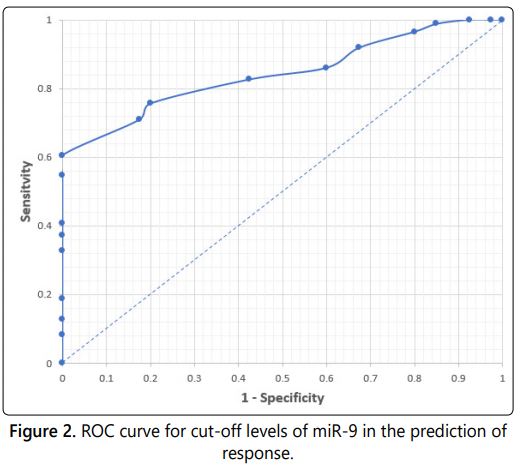
Receiver operating characteristic curves were plotted for each of miR-135 (Figure 1) and miR-9 (Figure 2) to identify the best cut-off point (Table 3). Regarding miR-135, a value of 27.59 was the best cut-off point to predict response. So, miR-135levels of 27.59 or lesser provided a sensitivity of 90.70%, a specificity of 90% and a positive predictive value of 95.12%, negative predictive value of 81.82 %. According to these results, miR-135 had a good predictive ability.
While in miR-9, a value of 27.59 was the best cut-off point to predict response. So, miR-9 levels of 27.59 or lesser provided a sensitivity of 75.58%, a specificity of 80.00% and a positive predictive value of 89.04%, negative predictive value of 60.38%.
Discussion
In Egypt, high frequency of breast cancer (48.8) followed by liver (24.4), brain (8.0), ovary (6.3) and non-Hodgkin lymphoma (6.1). Among females, the pattern in Lower, Middle, and Upper Egypt was dominated by the high frequency of breast cancer (33.8%, 26.8% and 38.7% resp.) and liver cancer (16.4%, 8.9% and 5.1% resp) [3].
One in eight women will be diagnosed with breast cancer in their lifetime. It is the second leading cause of death in women, with >40,000 deaths annually. Although the incidence and mortality rates of breast cancer in western countries have decreased or at least been stable over the last few decades, both rates are increasing in many developing countries [21].
Consequently, there is an urgent need for diagnostically sensitive, specific, and non invasive markers for early breast cancer detection. miRNAs could emerge as biomarkers for the diagnosis and prognosis of different diseases, including breast cancer as the expression patterns and levels of specific miRNAs could reflect altered physiological and pathological conditions, due to their high stability in human serum.
MicroRNAs are approximately 22-nucleotide, non-coding, endogenous RNA molecules that are over expressed in many human cancers [22].
miRNAs in the circulatory system can act as valuable diagnostic markers for early detection of BC [23]. miRNAs are associated with the regulation of oncogenes and tumor suppressor genes [19].
In the present study, we set out to analyze the expression patterns of miR-135and miR-9 as a single biomarker. Our results demonstrated that the expression levels of miR-135 and miR-9 are discriminate between breast cancer patients and healthy control subjects, with high specificity and sensitivity using ROC curve analysis and a fold change of 0.96 and 0.84 respectively.
The results showed that miR-135 has considerable diagnostic power in discriminating between breast cancer patients and control subjects, yielding an AUC of 0.96 with a sensitivity of 90.7% and a specificity of 90% and a positive predictive value of 95.12%, negative predictive value of 81.82%.
The result of miR-9, it has less diagnostic power than miR-135, yielding an AUC of 0.84 with a sensitivity of 75.6% and a specificity of 80% and positive predictive value of 89.04%, negative predictive value of 60.38% for the diagnosis of breast cancer. So, serum mi-R 135a can be used as a screening test to diagnose breast cancer as it showed high sensitivity. While other study showed in their ROC curve analysis that the best cut-off value for detection of cancer for miRNA-9 is 0.56 with sensitivity 75% and specificity 80% [24].
With regard to the clinicopathological features, both levels of miR135 and miR-9 showed no significant difference in age groups, BMI, different receptors and pathological types. However, levels of miR-135 showed statistical significant difference in tumor size groups, different grading and staging levels, while levels of miR-9 showed statistical significant differences among tumor size, lymph node groups and staging levels.
Our results showed significant up regulation of serum level of miR-135 in cases of breast cancer (mean value=214.7) compared to normal control (mean value=26.7), (p-value ≤ .00 1). These results are confirmed with the study that found miR-135a was up regulated in HCC cell lines and tissues when they examined its expression in HCC cell lines and clinical tissue [25]. However other study results showed that MiR-135a levels are significantly lower in glioblastoma cells compared to normal brain tissue [26].
Our results showed a significant relation between the gene expression of miRNA-135 in serum and tumor size (p=<0.01), different grades and staging levels. Meanwhile, there was no significant relation between the gene expression of miRNA-135 in serum and node state, metastatic state, Estrogen and Progesterone Receptor (ER/PR) state and tumor type in patients and this was not in agreement with the study examined expression levels of miR-135a in tissues and cells by poly (A)-RT PCR and found that miR-135a suppresses the expression of HOXA10 which induce p53 expression in breast cancer cells, and which results in increased migration and invasion [27].
Different mechanisms are involved in the progression of cancer through miR-9. miR-9 can promote metastasis by targeting leukemia inhibitory factor receptor to down-regulate Hippo signaling pathway [29]. It can also stimulate tumor angiogenesis, demonstrated that dysregulation of miR-9 can up-regulate vascular endothelial growth factor A, which is a proangiogenetic factor that works through β-catenin pathway by down-regulating E-cadherin [19].
The miR-9 can promote vascular sprouting in TNBC cell lines upon ligand-dependent stimulation of PDGFRβ signaling, and these mechanisms might attribute to TNBC progression [28]. Many studies have shown that miR-9 facilitates EMT and angiogenesis and suggested it as a possible oncomiR.
There are a few studies that have also demonstrated that miR-9 functions as a tumor suppressor in breast cancer by promoting apoptosis and inhibiting cell cycle [29]. Our results showed a significant relation between the gene expression of miRNA-9 in serum and tumor size (p=0.004), nodal status and staging levels, these results are confirmed with results who examined two different sets of breast cancer samples and evaluate 3 miRNAs (miR-200, miR-9, and miR-155) and found that miR-9 expression level was significantly higher in tumors with high T stage, high histologic grade [30].
There was no significant relation between the gene expression of miRNA-9 in serum and different grades, metastatic state, Estrogen and Progesterone Receptor (ER/PR) state and tumor type in patients.
Our results showed significant upregulation of serum level of miR-9 in cases of breast cancer (mean value=215) compared to normal control (mean value=25.3), (p-value ≤ .001). These results are confirmed with the study that investigated whether miR levels in breast tumors are predictive of breast cancer local recurrence (LR) by screening the expression levels of 754 human miRs using miR arrays they found eight candidate miRs that showed significant differences between tumors with and without LR were further verified using real-time PCR. Higher expression of miR-9 was significantly associated with breast cancer LR in all cases as well as the subset of estrogen receptor (ER) positive cases (p=0.02) [20,30].
Our results showed that miR-9 was more highly expressed in HER2+ and triple negative subtypes than in luminal A, luminal B, and luminal B/HER2+ subtypes (p=0.004, p=0.002, p=0.001, respectively). This finding is in consistence with several other reports implying that poor progression of the disease is associated with negative receptor proteins [31].
Conclusion
Our study establishes the role of miRNA 135 and miRNA 9 in breast cancer patientʼs disease progression and prognosis.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136(5): E359-E386. doi: 10.1002/ijc.29210
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017; 67(1): 7-30. doi: 10.3322/caac.21387
- Ibrahim AS, Khaled HM, Mikhail NHN, Baraka H, Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014; 437971. doi: 10.1155/2014/437971
- Ibrahim SS, Hafez EE, Hashishe MM. Presymptomatic breast cancer in Egypt: role of BRCA1 and BRCA2 tumor suppressor genes mutations detection. J Exp Clin Cancer Res. 2010; 29: 82. doi: 10.1186/1756-9966-29-82
- Williams NS, Bulstrode CJK, OʼConnel PR. Bailey and loveʼs short practice of surgery. 25th edition, Hodder Arnold. 2008.
- Chen Y, Zhang J, Wang H, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012; 12: 111. doi: 10.1186/1471-2407-12-111
- Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006: 6(4): 259-269. doi: 10.1038/nrc1840
- Xi JJ. MicroRNAs in Cancer. Cancer Treat Res. 2013; 158: 119-137. doi: 10.1007/978-3-642-31659-3_5
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009; 10(10): 704-714. doi: 10.1038/nrg2634
- Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007; 8(10): R214. doi: 10.1186/gb-2007-8-10-r214
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435(7049): 834-838. doi: 10.1038/nature03702
- Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008; 68(14): 5795-802. doi: 10.1158/0008-5472.CAN-08-0951
- Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008; 105(37): 13906-13911. doi: 10.1073/pnas.0804438105
- Pang RT, Liu WM, Leung CO, et al. miR-135A regulates preimplantation embryo development through down-regulation of E3 Ubiquitin Ligase Seven in Absentia Homolog 1A(SIAH1A) expression. PLoS One. 2011; 6(11): e27878. doi: 10.1371/journal.pone.0027878
- Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 Regulates HOXA10 Expression in Endometriosis. J Clin Endocrinol Metab. 2011; 96(12): E1925-1933. doi: 10.1210/jc.2011-1231
- Navarro A, Diaz T, Martinez A, et al. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood. 2009; 114(14): 2945-2951. doi: 10.1182/blood-2009-02-204842
- Liu S, Guo W, Shi J, et al. MicroRNA-135a contributes to the development of portal vein tumorthrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012; 56(2): 389-396. doi: 10.1016/j.jhep.2011.08.008
- Taipaleenmaki H, Browne G, Akech J, et al. Targeting of Runx2 by miR135 and miR-203 Impairs Progression of Breast Cancer and Metastatic Bone Disease. Cancer Res. 2015; 75(7): 1433-1444. doi: 10.1158/0008-5472.CAN-14
- Ma L, Young J, Prabhala H, et al. MiR-9, a MYC/MYCN activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010; 12(3): 247-256. doi: 10.1038/ncb2024
- Zhou X, Marian C, Makambi KH, et al. MicroRNA-9 as potential biomarker for breast cancer local recurrence and tumor estrogen receptor status. PLoS One. 2012; 7(6): e39011. doi: 10.1371/journal.pone.0039011
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61(2): 69-90. doi: 10.3322/caac.20107
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006; 20(5): 515-524. doi: 10.1101/gad.1399806
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008; 141(5): 672-675. doi: 10.1111/j.1365-2141.2008.07077.x
- Lyng MB, Kodahl AR, Binder H, Ditzel HJ. Prospective validation of a blood-based 9-miRNA profile for early detection of breast cancer in a cohort of women examined by clinical mammography. Mol Oncol. 2016; 10(10): 1621-1626. doi: 10.1016/j.molonc.2016.10.004
- Zeng YB, Liang XH, Zhang GX, et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016; 16(1): 63. doi: 10.1186/s12935-016-0328-z
- Gomez Zubieta DM, Hamood MA, Beydoun R, Pall AE, Kondapalli KC. MicroRNA-135a regulates NHE9 to inhibit proliferation and migration of glioblastoma cells. Cell Commun Signal. 2017; 15(1): 55. doi: 10.1186/s12964-017-0209-7
- Chen D, Sun Y, Wei Y, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012; 18(10): 1511-1517. doi: 10.1038/nm.2940
- DʼIppolito E, Plantamura I, Bongiovanni L, et al. miR-9 and miR-200 Regulate PDGFRβ-Mediated Endothelial Differentiation of Tumor Cells in Triple-Negative Breast Cancer. Cancer Res. 2016; 76(18): 5562-5572. doi: 10.1158/0008-5472.CAN-16-0140
- Selcuklu SD, Donoghue MT, Rehmet K, et al. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012; 287(35): 29516-29528. doi: 10.1074/jbc.M111.335943
- Gwak JM, Kim HJ, Kim EJ, et al. MicroRNA-9 is associated with epithelialmesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat. 2014; 147(1): 39-49. doi: 10.1007/s10549-014-3069-5
- Heneghan HM, Millar N, Lowery AJ, Sweeny KJ, Kerin MJ. MicroRNAs as Novel Biomarkers for Breast Cancer. J Oncol. 2010; 950201. doi: 10.1155/2010/950201


