Research Article
Study of P53 Gene Mutations as a New Early Diagnostic Markers of Hepatocellular Carcinoma in Egyptian Patients
1Department of Clinical Pathology, Tanta University Hospital, Egypt
2Department of Tropical Medicine, Tanta University Hospital, Egypt
*Corresponding author: Amal Helmy Abd Elhameed, Professor of Clinical Pathology Department, Tanta University Hospital, Egypt, Tel: 01001856164, E-mail: amalhelmy2015@gmail.com
Received: October 16, 2017 Accepted: January 15, 2018 Published: January 20, 2018
Citation: Abd Elhameed AH, Abo-Elenein AM, Ibrahim WS, El-kassas GM, Noweir MA. Study of P53 Gene Mutations as a New Early Diagnostic Markers of Hepatocellular Carcinoma in Egyptian Patients. Madridge J Oncogenesis. 2018; 2(1): 21-29. doi: 10.18689/mjo-1000104
Copyright: © 2018 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Background: Hepatocellular carcinoma (HCC) is the fifth most common malignancy in the world and the third most common cause of cancer related death worldwide. The outcome of this disease is poor as a result of late presentation. To improve the prognosis of this disease, several techniques have been advocated for screening and surveillance of high risk subjects, this is to facilitate early diagnosis at a stage when the disease is potentially curable. Recent researches have stated that p53 (exon 7) is a hotspot for point mutation in HCC and this mutation could be regarded as a new biomarker in HCC earlier diagnosis.
The aim: The aim of this work is to study p53 gene mutations (exon7) as a new early diagnostic marker of HCC in Egyptian patients.
Subjects and Methods: This study will be carried out on three groups: Group I HCC (40 patients), group IIcirrhosis (40 patients) and group III (20 controls). All groups will be subjected to history taking, clinical examination, abdominal ultasonography with or without triphasic CT scanning, routine laboratory investigations, alpha fetoprotien, hepatitis viral markers and detection of mutations in exon 7 of p53 gene by PCR-RFLEP followed by confirmatory direct DNA sequencing.
Results: In this study it was found that, there was no P53 mutations could be detected in the control group (GG), while in the HCC group, 12/40 (40%) patients showed G→T point mutation at codon 249 of exon 7, and in cirrhosis group 7/40 (17.5%) patients showed G→T point mutation. 8/40 (20%) of HCC patients and 6/40 (15%) of cirrhosis patients were heterozygous (GT), 4/40 (10%) of HCC patients and 1/40 (2.5%) of cirrhosis patients were homozygous (TT), while 28/40 (70%)of HCC patients and 33/40 (82.5)of cirrhosis patients were wild (GG).
Conclusion: P53 gene mutation (G to T codon 249) may be regarded as an early molecular marker in the detection of HCC in Egyptian patients.
Keywords: HCC; AFP; P53.
Introduction
HCC accounts for most primary cancers of the liver [1]. It represents an international public health problem and concern as one of the most common and deadly cancers [2]. Worldwide, it is the fifth most common cancer in men worldwide (7.9% of all cancers) and the seventh in women (6.5% of all cancers) and the third cause of cancer related mortalities [3]. It accounts 9.2% of all cancer deaths [3, 4].
Most of the burden of liver cancer is in developing countries, where almost 85% of the cases occur. HCC is the most common form of liver cancer; most cases of HCC (approximately 80%) are associated with chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infections. Variations in the age, sex and race of HCC rates in different geographic regions are likely to be related to differences in the prevalence of hepatitis viruses in the populations, as well as the timing of the spread of the viral infection and the age of individuals at the time of the infection [5]. Almost half of the data on HCC cases in Africa came from Egypt [6], being simultaneously plagued with the highest prevalence of HCV in the world [7]. While 30% of HCV infected individuals may clear the infection naturally, the remaining 70% will develop chronic disease that may result in liver cirrhosis and/or HCC [8].
Liver cancer exhibits no symptoms in the early stage, whereas clinical symptoms are evident in the advanced stage, leading to an unsatisfactory curative effect. In 2008, HCC was listed as the third most lethal cancer type. Early diagnosis of HCC and effective treatment are likely to prolong the lifetime of liver cancer patients. Thus, the identification of new high sensitivity and specificity markers for HCC are essential [9].
P53, also known as TP53 or tumor protein is a gene that codes for a protein that regulates the cell cycle and hence functions as a tumor suppression [10]. Tumor suppressor genes are normal genes that slow down cell division, repair DNA mistakes, and tell cells when to die (a process known as apoptosis). When tumor suppressor genes don't work properly, cells can grow out of control, which can lead to cancer. Tumor suppressor genes code for proteins that serve as the “stop” signals that tell the cell to leave the cell cycle and stop dividing [11].
It is very important for cells in multi cellular organisms to suppress cancer [12]. P53 has been described as “the guardian of the genome”, referring to its role in conserving stability by preventing genome mutation. The p53 tumor suppressor plays a key role in organizing cellular responses to various types of stresses, including DNA damage and oncogene activation with apoptosis, cell cycle arrest or DNA repair as shown in Figure (1) [13]. Malfunction and mutations of p53have been found in most of human cancers, leading to deregulated p53 activity that allows proliferating and surviving unchecked Figure (2)[14].
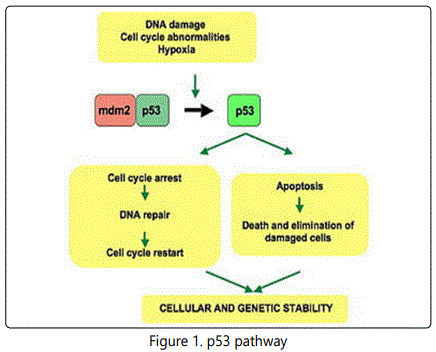
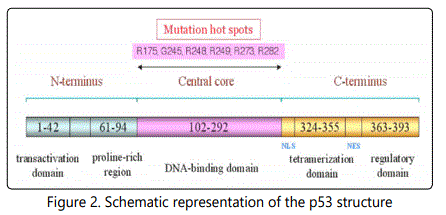
NLS, nuclear localization signal sequence; NES, nuclear export signal sequence.
The development of HCC is a multi-stage process and closely related to viral infection and chemical carcinogens. In the molecular aspect, dysregulation of growth factors, receptors and their downstream signaling pathway components represents a central pro-tumorigenic principle in human hepatocarcinogenesis [15]. p53 is one of the tumor relevant factors. Alteration of p53 status is an important intrinsic factor in this process. Its inactivation by point mutations within its gene or allelic deletions as well as the complex formation of wild (wt) p53 with several extrinsic factors, such as aflatoxin, HBV, nutrition, alcohol, and trace elements, are thought to initiate or/and promote the hepatocarcinogenesis, Figure 3 [16].
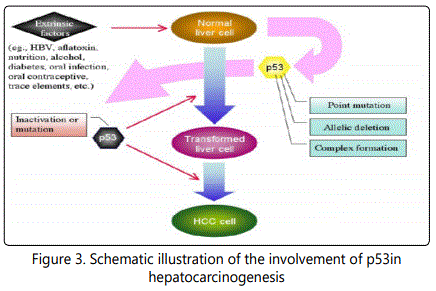
Aim of the Work
The aim of this work is to study p53 gene mutations (exon7) as a new early diagnostic marker of HCC in Egyptian patients.
Subjects and Method
After ethical committee approval from Research Center in Tanta University and written consent from all subjects, this current study was conducted on 100 Egyptian adult subjects, 80 patients with liver disease, selected from inpatient and outpatient of Tropical Medicine Department of Tanta University Hospital and 20age and sex matched healthy volunteers as a control group. The studied individuals were divided into 3 groups: Group I (HCC) included 40 patients of HCC that were diagnosed by clinical examination, radiological investigation, including abdominal U/S, triphasic CT and laboratory investigations. Group II (Cirrhosis): included 40 patients with liver cirrhosis without HCC, they were diagnosed by U/S findings and biochemical evidence of parenchymal damage. Group III (control): included 20 apparently healthy volunteers, they were seronegative for HBs markers and HCV Ab and with normal liver function test.
After written consentall patients and controls were subjected to the following:Full history taking, thorough clinical examination, abdominal ultrasonography, triphasic C.T (for patients only).Routine laboratory investigations (liver function tests including: AL, AST, serum albumin, total and direct bilirubin, prothrombin time and INR, complete blood count, serum urea and creatinine, fasting and 2-hours post prandial blood glucose levels, serum AFP, hepatitis markers (HCV-Abs, HBs Ag) and detection of mutations in exon 7 of p53 gene by PCR-RFLEP followed by confirmatory direct DNA sequencing.
The analysis of liver functions, fasting and 2-hours post prandial blood glucose levels, serum urea and creatinine were performed on Konelab Thermo Scientific (Thermo Scientific – Finland) autoanalyzer. Hepatitis markers (HCV-Abs and HBs Ag) were done by ELISA and confirmed by quantitative PCR for positive cases. HCV antibody was done by anti HCV ELISA kit which is provided by WKEA MED SUPPLIES CORP (www. wkeamedsupplies. com). HBsAg was done by Hepatitis B Surface Antigen (HBsAg) ELISA Kit which is provided by Prechek Bio Inc Company USA (www.prechecbio.com).Full blood count using ERMA PCE-210 cell counter.Serum alpha fetoprotein was estimated by a direct solid phase sandwich assay based on the ELISA, supplied by Assay Pro LLC. Human (α-fetoproteinELISA Kit, catalogue number EF6011).
Identification of p53 gene mutations: DNA analysis was carried out for all studied groups (HCC, cirrhosis and control).
DNA extraction protocol
DNA was isolated from the peripheral whole blood by using DNA extraction kits (QIAamp DNA Blood Mini Kits). The concentration of DNA was measured by ultraviolet spectrophotometer at 260 and 280 wave length and ratio of 260/280 was detected. Pure DNA had an A260/A280 ratio of (1.7- 2.0), the extracted DNA was stored at -20© until further processing.
Conventional Polymerase Chain Reaction (PCR)
DNA extracts were amplified for the P53 gene using the primers
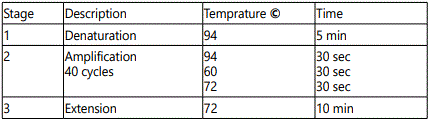
F: 5′-CTTGCCACAGGTCTCCCCAA-3′ and R:5′-AGGGGTCAGCGGCAAGCAGA-3′. The 25 µl reaction included 18.3 µl ddH2O, 2.5 µl 10 × buffer, 1 µl dNTPs, 0.5 µl primer 1, 0.5 µl primer 2, 0.2 µl Taq DNA polymerase, 2 µl DNA templates. (Roche Molecular Biochemicals). The samples were mainly, [denaturated at 94C for 5min, amplification 40 cycles (94 C for 30 seconds, 60 C for 35 sec,72 C for 30 sec) followed by final extension at 72 C for 10 min ]. Visualization of amplified material by gel electrophoresis apparatus (Biometra, Germany).The amplification products (254 bp) were visualized by staining with ethidium bromide, after electrophoresis on 2 % agarose gel.
Detection of P53 gene mutation (codon 249) using restriction enzyme fragment length polymorphism
The test samples were digested with restriction enzyme HaeIII Enzyme (provided by Fermentas Life Science, lot no 00011400.
The reaction was as follows:
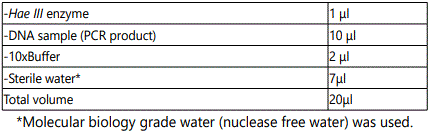
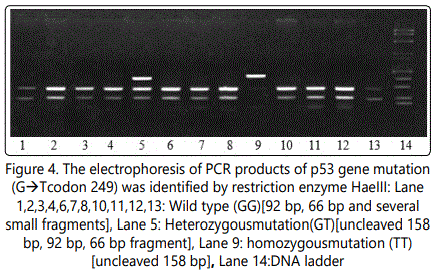
After preparing the reaction tubes, the components were mixed by gentle pipetting and tapping. The tubes were incubated at 37 Co for 18 hours (over night).After incubation, 1/10 volume of 10x loading buffer was added to aliquate of the reaction mixture. The samples were run on agarose gel. Wild type detected as 92 bp, 66 bp and several small fragments, heterozygous mutation detected as uncleaved 158 bp, 92 bp, 66 bp, while homozygous mutation detected as uncleaved 158 bp.
Purification of PCR product
After visualization PCR band by gel electrophoresis, the amplified PCR product of each sample was purified using QIA quick PCR purification kit.
DNA concentration and purity: Pure DNA had an A 260/ A280 ratio of 1.7-2.0.
Cycle sequencing: [17]
Preparation of the cycle sequence reaction mixture for the gene was done as the following:

The cycle sequence was done in the thermal cycler (Biometra) programmed as:
Purification of DNA following cycle sequencing
The amplified PCR product of each sample was purified using QIA quick PCR purification kit.
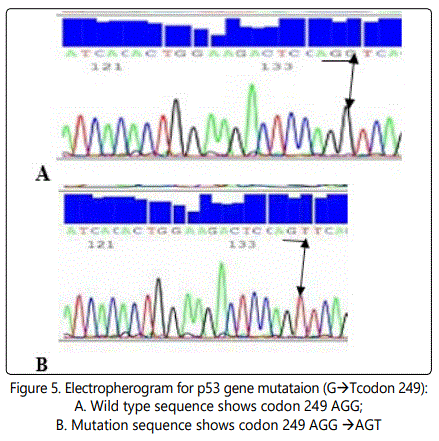
DNA sequencing: Injection of purified cycle sequence reaction into automated genetic analyzer (Applied Biosystem 310 Genetic Analyzer, Germany).Then comparing the wild type of P53 gene in healthy control with the mutated allele of the patients and also comparing all our results of DNA sequencing for all patients and controls with the reference gene sequence on the gene bank.
Statistical Analysis
Statistical presentation and analysis of the present study was conducted, using the mean, standard deviation and chisquare test by SPSS V.16.
Results
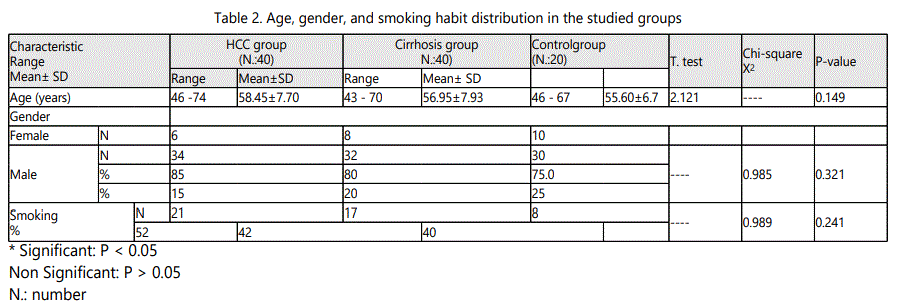
In HCC group the age ranged from 46 to 74 years with a mean value of 58.45 ± 7.70, in cirrhosis group it ranged from 43 to 70 year with a mean value of 56.95 ± 7.93, while in control group it ranged from 46 to 67 year with a mean value of 55.60 ± 6.7.As regard sex, in HCC group, male patients were 34 (85 %) and females patients were 6 (15.0%), in cirrhosis group they were 32 (80 %) males and 8 (20.0%) females, while in control group they were 30 (75.0 %) males and 10 (25.0%) females. It was proved no significant difference in both age and sex among all studied groups (p > 0.05).
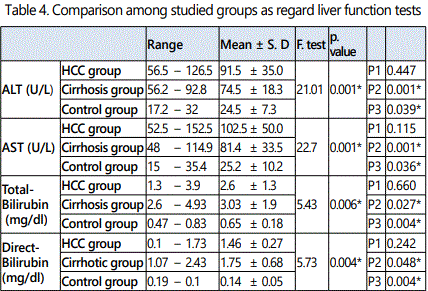
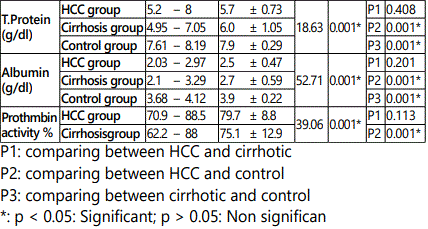
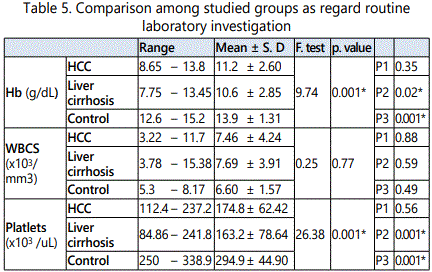
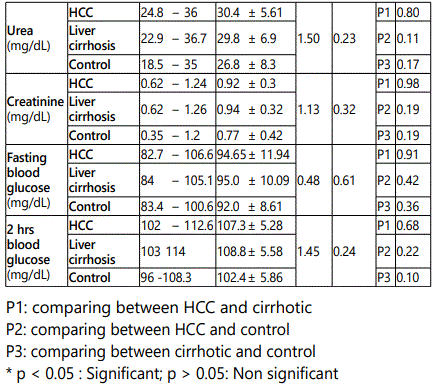
As regard HCV-Ab positive cases they were 30/40 (75.0 %), 32/40 (80.0%) and 0/20 (0%) in HCC, cirrhosis and control groups respectively. There was a statistically significant increase in the percentages of HCV-Ab positive in HCC and cirrhosis group when compared to control group (p < 0.05), while there was no statistically significant difference in HCC group when compared to cirrhosis group (p > 0.05). As regard HBs Ag positive cases they were 6/40 (15 %), 8/20 (20.0 %), and 0/20 (0%) in HCC, cirrhosis and control groups respectively. There was no statistically significant difference in the percentages of HBs Ag among studied groups (p > 0.05).
In HCC group AFP ranged from 2.6 to 2200 ng/dl with a mean value of 273.89 ± 131.85, in cirrhosis group it ranged from 2.0 to 76.5 ng/dlDemonstrate the sexfeto-proteinrom 17-40 md/dl wit with a mean value of 35.96 ± 15.65, while in control group it ranged from 3.0 to 5.2 ng/dl with a mean value of 4.06 ±0.62. There was a statistically significant increase in the mean value of AFP in HCC group when compared to cirrhosis and control group, as well as in its mean value in cirrhosis group when compared to control group (p <0.05).
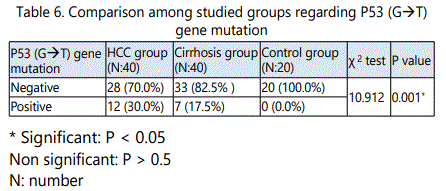
As regard p53G→T gene mutation, 12/40 (30.0%), 7/40 (17.5%) and 0/20 (0.0%) were positive in HCC, cirrhosis and control groups respectively. In HCC group, 28 (70%)were wild type (GG),8 (66.7%)were heterozygous (GT), while 4 (33.3%) were homozygous (TT). In cirrhosis group33 (82.5)were wild type (GG), 6 (85.7%)were heterozygous (GT) and1 (14.3)was homozygous (TT).
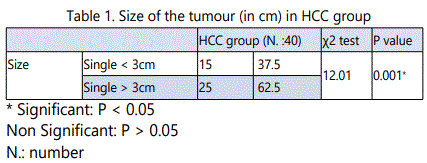
In HCC patients with tumour size<3cm, the mean value of serum AFP was 145.85 ± 73.87, while in HCC patients with tumoursize> 3cm, its mean valuewas541.76±175.8. In HCC patients, with tumor size <3 cm, 2/15 patients (5 %) showed p53 (G→T) gene mutation, while in HCC patients with tumor size >3 cm, 10/25 patients (25 %) showed p53 (G→T) gene mutation. There were a statistically significant increase in the mean values of serum AFP as well as in the pecentage of P53 (G → T) gene mutation in HCC patients with the tumour size > 3cm when compared to patients with the tumour size< 3cm (p<0.05).
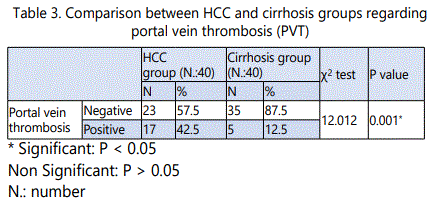
The mean values of serum AFP in HCC patients with negative PVT were 186.43± 91.85, while in patients with PVT were 267.14± 130.24. In HCC group with negative PVT 7/23 patients (17.5 %) showed p53 gene(G→T) mutation, while in HCC group with PVT5/17 patients (12.5%) showed p53 gene mutation. There were no statistically significant difference in the mean value of serum AFP as well as in the percentage of P53(G→T) gene mutation in HCC patients with and without PVT (p >0.05).
Discussion
HCC is the most common primary liver malignancy, the fourth leading cause of cancer and the third leading cause of cancer deaths worldwide. The disease is predominant in Asia and Africa, but its incidence is steadily increasing throughout the rest of the world. Most HCC develop in patients with a history of chronic hepatitis or cirrhosis in which there is continuous inflammation and regeneration of hepatocytes [18-21].
In Egypt, incidence of HCC is currently increasing, which may be the result of a shift in the relative importance of HBV and HCV as primary risk factors. Egypt has possibly the highest HCV prevalence worldwide. In Egypt, HCC is the second most common cancer in men and the 6th most common cancers in women [22]. It is estimated that the problem of HCC will increase until it reaches its peak in the year 2018 [23].
Several studies have reported that alterations of the p53 gene are correlated with tumor differentiation, vascular invasion, and tumor stage in HCC. Moreover, aberrations of the p53 gene have been shown to be prognostic indicators associated with recurrence-free survival and overall survival in HCC patients. P53 may open new avenues for the prevention, early detection and treatment of HCC through the development of targeted therapies [24-27].
The aim of this work is to study p53 gene mutations (exon7) as a new early diagnostic marker of hepatocellular carcinoma in Egyptian patients. From our study it was clear that, HCC is more common in male gender than female. These results are nearly in accordance to those reported for Egyptian patients by Ashraf et al., (2014) [28]. Men are more likely to be affected than women with HCC. The reasons are not well understood but several factors may explain that, males are more likely to be infected with HBV and HCV as well as, cigarette smoking and alcohol consumption are more common in men. Women estrogen inhibit interleukin 6 which reduce hepatic damage. Testosterone has been shown to correlate with HCC indicating a probable role for the sex hormones in the development of HCC [29].
The present study showed that there was a significant increase in the mean value of AFP in HCC group when compared to cirrhosis and control group, as well as in cirrhosis group when compared to control group. This was in agreement with several studies that found AFP was significantly increased in HCC patients when compared to cirrhotic patients and healthy control [30-32]. El Shafie et al., (2012) also reported that the serum levels of AFP are significantly elevated in chronic liver diseases and more elevated in HCC cases [33]. However, Shaheen et al., (2015) reported that not all tumors secrete AFP, and serum concentrations are normal in up to 40 % of small HCC. Likewise, it was reported that only 10-20% of patients in early stages of HCC present with abnormal AFP serum levels (Yamashita et al., 2008) [34,35].
AFP together with iconography and pathology detection are commonly used in clinical early diagnosis for liver cancer, however, it does not yield satisfactory results in the early diagnosis of HCC, particularly AFP-negative HCC. These false-negative results limit the universality of its application. In recent years, the development of molecular biology has led to the successful exploration and identification of markers for HCC, which is expected to improve the early diagnostic rate, treatment effect in addition to curative satisfaction [36].
Serum AFP is the golden standard of diagnostic markers for hepatocellular carcinoma. However; its diagnostic value is more and more questioned. Serum AFP is only observed in about 60%-70% of HCC patients and, to a lesser extent (33-65%), in patients with smaller HCCs. Moreover, the non-specific elevation of serum AFP in 11%-47% of patients with liver cirrhosis have led to the exclusion of AFP as a screening test alone for HCC patients from the European Association for the Study of the liver (EASL) guidelines [23]. The Practice Guidelines of the American Association for the Study of Liver Diseases (AASLD) July 2010 also rejected AFP alone whether for the surveillance or the diagnosis of HCC. This highlights the need to identify new serologic biomarkers that would be minimally-invasive, simple and reliable for the early detection of HCC [37,38].
In this study it was found that, there was no P53 mutation should be detected in the control group (GG), while in the HCC group, 12/40 (40%), and in cirrhosis group 7/40 (17.5%) showed G→T point mutation. 8/40 (20%) of HCC patients and 6/40 (15%) of cirrhosis patients were Heterozygous (GT), 4/40 (10%) of HCC patients and 1/40 (2.5%) of cirrhosis patients were Homozygous (TT),while 28/40 (70%)of HCC patients and 33/40 (82.5)of cirrhosis patients were wild (GG).
P53 is mutated in more than 50% of aflatoxin B1-induced HCC, in up to 45% of HBV-related HCC and in 13% of HCVrelated HCC [27, 39]. The mutations have been found in many sites of the P53 gene in cancers, resulting from a guanine to thymine (G toT) transversion at the third position of codon 249 of the exon 7 of the gene,it is the particular hotspot mutation found almost exclusively in patients with HCC. Approximately 50% of patients with HCC have this p53 hotspot mutation [40-42]. Thus, the P53 codon 249 hotspot mutation could possibly be a DNA marker for HCC screening [43].
Igete et al., (2008) reported that the prevalence of the p53 codon 249 mutation from plasma DNA of hepatocellular carcinoma patients is significantly higher than among controls in South-Western Nigeria and the presence of this mutation is significantly associated with HCC in this region [44]. Also, the results of several studies has shown that codon 249 is one of the most important sites introduced as a hot-spot for p53 gene in HCC [41, 45, 46].
Kirk et al., (2000) also reported the presence of codon 249 p53 mutations in plasma DNA isolated from the plasma of individuals from a West African population at high risk for HCC. This “hotspot” mutation was detected in 36% of patients with HCC, findings that may lead to the earlier detection of HCC and should facilitate further molecular epidemiologic studies on the development of HCC [47].
Selena et al. (2011) reported that eight of nine high molecular weight urine DNA fractions of HCC urine samples contained a detectable p53 mutation, while did not detect any p53 mutation in normal urine, which suggests that the qualitative assay for TP53 codon 249 might be sufficient to use in a urine or blood screening test for HCC[43].
In contrast to the current study, El-Dein et al., (2010) reported that the incidence of point of mutation of p53 at codon 249 mutations in exon 7 may not play any role in carcinogenesis of HCC [48]. Huang et al., (2003) found that the codon 249 in p53 mutation in plasma is strongly associated with hepatocellular carcinoma in Qidong patients and it was also detected in plasma DNA of Qidong cirrhotic and healthy controls; although it was at a much lower frequency. These results recommended that codon 249 p53 mutation should be used as a new early diagnostic marker for HCC [49]. These contradictory results may be explained by different sample size, population difference, or difference in the invironmental factors that trigger the genetic mutations as exposure to aflatoxins, and this directed us for the need to clarify this point in HCC in Egyptian patients.
Jackson et al., (2001) also reported that a high percentage of the tumors from Qidong had G to T mutation at the third base of codon 249 of the p53 gene than tumors from Shanghai (46.7% compared with 30%). The mutation frequency corresponds to exposure to aflatoxins because these areas have high and intermediate exposure levels, respectively. DNA circulating in the serum or plasma can be successfully retrieved and used as surrogate material to analyze for genetic alterations present in the original tumor [50].
Although the mechanisms accounting the presence of this circulating DNA are uncertain, there is some evidence that the DNA are released from the tumor as a glyconucleoprotein complex, and may protect the DNA from degradation by nucleases. Although the relationship between the release of tumor DNA into the plasma and necrosis of the tumor, apoptosis or other selective cellular processes is not understood, p53 gene in plasma or serum may be used as a source of tumor-specific DNA (Huang et al., 2003) [49].
In this study, there was a statistically significant increase in the mean values of serum AFP in HCC patients with the tumour size > 3cm when compared to patients with the tumour < 3cm. In contrast, there was no statistically significant difference in mean value of serum AFP in patients with PVT when compared to patients with negativePVT. As regard P53 G to T gene mutation, it was a statistically significant increased in HCC patients with the tumour size > 3cm when compared to patients with the tumour < 3cm. Tumor size has been emphasized as one of the significant prognostic factors because of vascular invasion which may increasingly develop as the tumor grows. Other study showed that no significant correlation between p53 positivity and tumor size was found [51]. P53 has higher expression level in early and advanced stage HCC, compared to hepatitis and cirrhosis. Comparing the positive rate in early and advanced stage HCC, P53 is significantly higher in advanced stage HCC, implying P53 is one of the changing factors in advanced HCC [52, 53].
P53 pathway is a key responder to inflammatory stress. It responds to lower levels of DNA damage by cell cycle checkpoint arrest, facilitating DNA repair as an adapter in the formation of DNA repair protein complexes and transcriptional transactivation of DNA repair genes and to extensive DNA damage by mediating cell death [54].
Chronic inflammation and the generation of an excess reactive oxygen species, which causes oxidative DNA damage or increases susceptibility of somatic mutations, may be responsible for the larger proportion of P53 mutations accumulating in HCV-related cancer. Free radicals have been demonstrated to directly damage DNA and proteins [53]. Tumour-specific mutations of the p53 gene often lead to synthesis of a faulty protein that has lost its normal growth regulatory functions. The mutation of the p53 gene in HCC suggests that loss of normal gene function may be a key step during malignant transformation of hepatocytes [34].
Sheen et al., (2003) considered the status of p53 mutations to be useful as a predictor affecting both the recurrence after resection and the prognosis after recurrence, even before the pathologic findings of recurrent HCCs are known. Therefore, it is important in the follow-up of patients after resection of HCCs. Thus, patients with p53 mutations have a worse prognosis than patients without such mutations, including survival after recurrence. Its significance was even greater than that of factors such as tumor size and vascular invasion. So it is recommended to study other p53 mutations including the other known hot-spot mutations, moreover, p53 mutational studies should be done on HCC from patients at various stages of the disease, early as well as metastatic phase.
We can draw a conclusion from these data that p53 mutation frequency of these patients groups (HCC and cirrhosis) provided a strong relationship between cirrhosis and the future development of HCC. So the detection of p53 gene mutataions may be useful as a maker of neoplastic development in those patients.So P53 gene mutation (G to T codon 249) may be regarded as an early molecular marker in the detection of HCC in Egyptian patients.
Acknowledgements
To Clinical Pathology Department, Faculty of Medicine, Egypt.
Conflict of Interest
The authors declare that they have no competing interests.
References