Research Article
Epithelial Mesenchymal Transition enhances the Chemosensitivity of Triple Negative Breast Cancer Cells to JQ1
1Research Assistant, Department of Biology and Environmental Sciences, University of New Haven, West Haven, USA
2Associate Dean and Professor, Department of Biology and Environmental Sciences, University of New Haven, West Haven, USA
*Corresponding author: Michael J Rossi, Associate Dean, University of New Haven, 300 Boston Post Road, West Haven, CT, 06516, USA, E-mail: MRossi@newhaven.edu
Received: October 25, 2017 Accepted: December 6, 2017 Published: December 12, 2017
Citation: You Y, Rossi MJ. Epithelial Mesenchymal Transition enhances the Chemosensitivity of Triple Negative Breast Cancer Cells to JQ1. Madridge J Oncogenesis. 2017; 1(1): 12-20. doi: 10.18689/mjo-1000103
Copyright: © 2017 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Triple negative breast cancers (TNBC) are particularly aggressive and more likely to recur than other subtypes of breast cancer. Bromodomain and extra terminal domain (BET) inhibitors have shown efficacy in several cancer models by inhibiting BET function. JQ1, a specific Bromodomain inhibitor, functions as a competitive inhibitor, with IC50 values from 92-112 nM.
Objectives
To study the inhibitory effect of JQ1 to TNBC cells in 2D and 3D culture systems.
Methods
In this study, triple negative mesenchymal-like cell lines (MDA-MB-231, Hs578T) and basal-like cell line (Hcc1143) were used to examine the inhibitory effect of JQ1. A 3D culture system was used to potentially activate an epithelial-mesenchymal transition (EMT) in these lines. MTT assays and 3D apoptosis assays were performed to determine the inhibitory effect of JQ1 to the TNBC cells.
Results
Markers for EMT were expressed when Hcc1143 cells were cultured in the 3D culture, but this was not seen for MDA-MB-231 and Hs578T cells. However all three cell lines had significantly higher bromodomain protein Brd4 mRNA expression in 3D cultures and increased the expression of bromodomain proteins compared to 2D cultures. Viability assays determined that the IC50 for JQ1 in the 2D system ranged from 30 to 170 nM. In the 3D system the IC50 ranged from 10 to 80 nM. Hcc1143 cells were more sensitive to JQ1 in the 3D system than in a 2D system.
Conclusion
JQ1 inhibited the growth of TNBC cells in both systems and shows potential for further studies of triple negative breast cancer.
Keywords: Triple Negative Breast Cancer; Epithelial-Mesenchymal Transition; Bromodomain Protein; JQ1.
Introduction
Breast cancer is the second most common cancer among American women, about 12% of women in the US will develop invasive breast cancer during their lives [1]. In the United States, The American Cancer Society estimated 255,180 new cases of invasive breast cancer and 40,610 deaths in 2017 [1]. Breast cancer is classified based on molecular properties including the expression of estrogen receptor (ERα), progesterone receptor (PgR) and human epidermal growth factor receptor 2 (HER2) [2]. Triple-negative breast cancers (TNBC) do not express the genes for ER, PgR or Her2. TNBC occurs in 10 - 20% of diagnosed breast cancers, but leads to 25% of all breast cancer deaths [3]. Depending on the stage of its diagnosis, triple-negative breast cancer can be particularly aggressive, and more likely to recur than other subtypes of breast cancer [2]. While triple-negative tumors generally do not respond to receptor targeted treatments, TNBC is typically responsive to initial chemotherapy [2].
Alsarraj and Hunter (2011) determined that bromodomaincontaining protein 4 (BRD4) functions as an inherited susceptibility gene for breast cancer progression and metastasis. They hypothesized that it could be a potential therapeutic target for TNBCs [4].
BRD4 is one member of Bromodomain and extraterminal domain family (BET), including BRD2, BRD3, and BRDT. BET protein domains were originally found as proteins associated with chromatin organization and nearly all nuclear histone acetyltransferases [5]. Proteins of the BET family are key mediators for the assembly of the positive transcription elongation factor b complex (P-TEFb), an event required for the initiation of transcription elongation [6]. The P-TEFb core complex, composed of cyclin-dependent kinase-9 and its activator cyclin T, functions to activate RNA polymerase II [7]. BET proteins are also required for maintaining a proper higher-order chromatin structure. Importantly, BET proteins have been shown to be implicated in cancer, where they can be expressed as malignant oncogenic fusion proteins together with nuclear protein in testis (NUT), resulting in very aggressive and poorly differentiated carcinomas that originate mainly from midline locations such as the head, neck, or mediastinum [8, 9].
JQ-1 is one of the bromodomain inhibitors specific for the BET family, competitively binding to the acetyl-lysine recognition pocket of BET bromodomains [9,10]. Inhibition of the BET bromodomain with JQ-1 demonstrated potent anticancer effects both in vitro and in vivo in several different hematopoietic cancers as well as NUT midline carcinoma [11,12].
In humans, cells reside in an extracellular matrix (ECM) consisting of a complex 3D fibrous meshwork with a wide distribution of fibers and peptidoglycans that provide complex biochemical and physical signals [13]. Commonly, a 3D cell culture system provides the culture environments where cell are allowed to interact with their surrounding cells in all three dimensions. Additionally, each type of cell is embedded in a specific 3D microenvironment [13]. Therefore, regular 2D cell culture systems are inadequate representations of the real living microenvironment, which often makes them unreliable predictors of in vivo drug efficacy and toxicity [14].
The development of cancer is a multistep process and the activation of epithelial-mesenchymal transition (EMT) plays a important role in cell dissemination [15,16]. EMT has also been shown to occur in wound healing, in organ fibrosis, and in the initiation of metastasis for cancer progression [17]. Whether the BRD2, BRD3 and BRD4 in TNBCs cultured in 2D and 3D will have similar gene and protein expression is not clear. To gain insights into the bromodomain protein's function on cell growth, the gene expression and protein expression of the BRD family were measured in TNBCs cultured in 2D and 3D conditions. We were also curious if MET and EMT will occur when TNBCs were growing in 3D systems compared to 2D systems. To answer these questions, cell proliferation in the presence of JQ1 was studied to clarify whether JQ1 inhibits TNBC growth. Cell viability assays were performed to determine the JQ1 inhibition of TNBC cells in 2D and 3D. The hypothesis was the 3D culture system would provide a more representative system to determine the effect of JQ1 on TNBC growth and viability.
Methods and Materials
Cell lines and culture conditions
The cell line MDA-MB-231 (ATCC® HTB-26™) was cultured in DMEM-F12 medium supplemented with 10% (v/v) fetal bovine serum (Serum Source International) and 1% (v/v) L-Glutamine with antibiotics (Sigma, USA). The cell line Hs578T cells (ATCC®HTB-126™) was cultured in DMEM medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) L-Glutamine with antibiotics. The cell line HCC143 (ATCC® CRL2321™) was cultured in RPMI-1260 medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) L-Glutamine with antibiotics. The cultures was incubated at 37°C and 5% CO2, maintained in 75 cm2 polystyrene coated tissue culture flasks and passaged every 3 days at 70% confluence.
3D cell Culture conditions
In 3D cell culture system, 3 ml of 10% (g/v) gelatin (Type A, Sigma, USA) was incubated in 100 mm cell culture dishes (Corning, Sigma, USA) for 30 minutes until the gelatin became solid. Then 106 cells were transferred and cultured with 6 ml growth medium for 48 hours.
Reverse transcription PCR
RNA extractions from cells cultured in 2D and 3D were performed by using the Trizol Kit (Ambion®, Life Technologies. USA). RT-PCR was performed using the Verso™ cDNA Kit (Thermo Scientific, AB-1453). cDNA was reverse transcribed from 1 μg RNA. PCR amplification was done under the following conditions: 47°C for 1 hour for cDNA synthesis and 95°C for 2 minutes for inactivation and hold at 4°C. The cDNA was diluted to 5 ng/µl.
Quantitative PCR
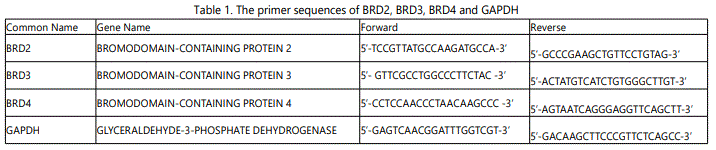
Quantitative PCR was performed using a reaction mix (PCR Kappa Supermix, Thermo Fisher, USA) containing Tag DNA polymerase, Tris-HCl, potassium chloride, magnesium chloride and deoxynucleoside triphosphates. In detail, 12.5 ng of cDNA was mixed with 5 µl of Supermix (Thermo Fisher, USA) and 2.5 μl of primer pairs (1 µM forward primer: 1 µM reverse primer). qPCR was performed in the Bio-Rad CFX96™ Real-Time System for 2 hours under the following conditions: 95°C for 15 minutes for initial melting; 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute for a total of 40 cycles; and 72°C for 10 minutes for final extension. Primers used are shown in Table 1.
RT-qPCR quantification
Cq values were measured by CFX Manager™ Software (Bio-Rad, USA). The average of Cq values of each gene was calculated using Excel software. The ΔΔCq method were performed to quantify the gene expression level, and GAPDH house keeping gene was used as controls for both 2D and 3D cultures.
Protein Extraction
500 μl of ice cold 1X cell lysis buffer (Cell Signaling, #9803, USA) was added into the collected cells and then incubated for 30 minutes on ice. After the centrifuge for 10 minutes at 14,000 xg at 4°C, the supernatant were collected for use.
Western Blot
For western blots, 40 mug total protein extraction lysates were loaded in 7.5% SDS-polyacrylamide gel and transferred to PVDF membranes. The membranes were blocked for 1 hour with the 5% milk/PBST (8 mM Na2HPO4, 150 mM NaCl, 2 mM KH2PO4, 3 mM KCl, 0.05% Tween® 20, pH 7.4) and then incubated with primary antibodies overnight separately: mouse anti-human BRD3 (2088C3a, Santa Cruz, USA, 1:1000); rabbit anti-human BRD4 (ab128874, Abcam, USA, 1:1000); rabbit anti-human Vimentin (5741, Cell Signaling, USA, 1:1000); rabbit anti-human YY1 (ab12132, Abcam, USA, 1:1000); rabbit anti-human Actin (ab8227, Abcam, USA, 1:2000) and mouse anti-human GAPDH (ab9483, Abcam, USA,1:10000). After incubation with the corresponding secondary antibody (goat anti-rabbit IgG (H+L) horseradish peroxidase (HRP) conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc., USA)) or anti-mouse IgG (H+L) horseradish peroxidase (HRP) conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc., USA) at a dilution of 1:10,000 for 2 hours, membranes were finally incubated with a chemiluminescent reagent (Clarity™ Western ECL Blotting Substrate, Bio Rad, USA) for 3 minutes and the chemiluminescent blots were recorded by FluorChem™ E System (ProteinSimple, USA).
Quantification Analysis of Western Blots
Western images were subsequently imaged with the Fluor Chem™ E System using the white light conversion screen and the silver stain (visible stain) application. The band analysis tools of Alpha View™ software were used to select and determine the background-subtracted density of the bands in all the gels and blots. Quantification analysis of Brd3 and Brd4 protein expressions was performed by using GAPDH as control for total protein.
2D Cell Viability Assay
MTT assays were performed to determine cell viability under the treatment of JQ1 in a 2D cell culture system. For this assay, 1x106 cells were transferred into each well of 48- well plates with 500 μl medium. Cells were treated with 1 μl JQ-1 and Carboplatin at various final concentrations (0 µM, 2.5 µM, 5 µM, 10 µM, 20 µM, 40 µM, 80 µM, 160 µM, 320 µM and 640 µM). After 3 days, 50 µl of 5 mg/ml MTT solution was directly added to all wells except the blank wells. After the incubation of 3 hours at 37°C, the medium in each well was removed and 500 µl of DMSO and 25 µl of Sorensen's glycine buffer (0.1 M glycine, 0.1 M NaCl equilibrated to pH 10.5 with 0.1 M NaOH) was added into each well. With gentle shaking, color was allowed to develop for at least 10 min at room temperature, and then the absorbance at 570 nm was determined. The ED50 of JQ-1 and Carboplatin for each cell line were determined by analyzing the data of three experiments using the Prism 6 Program (GraphPad Software, Inc, USA).
3D Live Cell Imaging
Effect of JQ1 on cell proliferation was investigated by recording time-lapse images of the cells for 3 days. 50 μl of 10% gelatin were plated in the 96 well plate and incubated for 30 minutes. MDA-MB-231 cells, Hs578T cells and Hcc1143 cells (6 × 103) were seeded into 96-well plates and allowed to attach for 24 hr. The media was replaced with fresh medium containing different concentrations of JQ1 (0 µM, 2.5 µM, 5 µM, 10 µM, 20 µM, 40 µM, 80 µM, 160 µM, 320 µM, 640 µM and 1280 µM). The plate was imaged in phase using an IncuCyte ZOOM™ Kinetic Imaging System (Essen Bioscience) every 2 hours for 3 days. Inbuilt software was used for analysis of the images to generate confluence data.
3D Cell Viability Assay
Cell Titer-Glo® 3D Cell Viability Assay (Promega, USA) were performed to determine cell viability under the treatment of JQ1 at 3D cell culture system. For this assay, 50 μl of 10% gelatin were plated into each well of the 96 well plate and incubated for 30 minutes. And then 6,000 cells were transferred into each well of 48-well plates with 100 μl medium. After 24 hours' culture, cells were treated with 1 μl of JQ-1 at various final concentrations (0 µM, 2.5 µM, 5 µM, 10 µM, 20 µM, 40 µM, 80 µM, 160 µM, 320 µM and 640 µM and 1280 µM). After 3 days, 3D cell viability assay were performed. In briefly, Cell Titer-Glo® 3D Reagent were thaw at 4°C overnight and placed in a 22°C water bath for 30 minutes prior to use. In 96 well plate, 50 ul of media was carefully took away from each well. 100 ul of 3D Reagent were added into the contents and incubated at room temperature for 30 minutes. Luminescence measurements were recorded by reading all wavelengths with SpectraMax Plus 384 Microplate Reader (Molecular Devices, California, USA). The IC50 of JQ-1 for each cell line were determined by analyzing the data of one experiment using the Prims 6 ™Program (GraphPad Software, Inc, USA).
Statistical analysis
The differences between two groups (n=3) were determined with Mann-Whitney test by using Graph Pad Prism 6 statistical software (La Jolla, CA). A p-value of ≤0.05 was considered as significant and data is presented as mean ± standard deviation of the mean.
Results
Gene expression of bromodomain proteins were induced in 3D cultures
A comparison of Cq values of BRD2, BRD3 and BRD4 gene expression from 2D and 3D cultures determined the effect of the 3D cell culture on the gene expression of BET family transcripts. GAPDH was used as control in this qPCR assay. All three TNBC cell lines had significantly higher BRD4 expression in 3D than in 2D at 24 hours (Figure 1). However, only Hcc1143 had significantly higher BRD2 and BRD3 expression in 3D compared to 2D cultures at 24 hours.
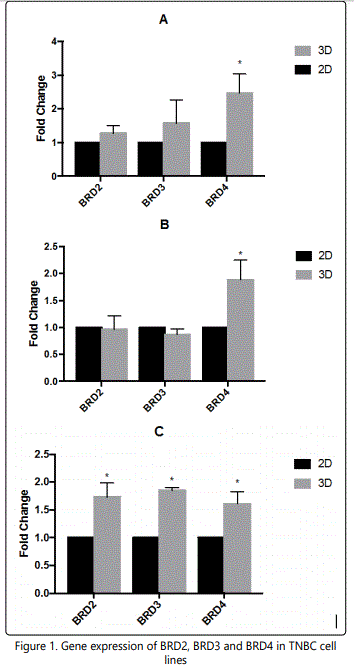
QPCR was performed using GAPDH as control. Gene expression of BRD2, BRD3 and BRD4. BRD4, determined by the ΔΔCq method, was significantly higher in 3D cultures for (A) MDA_MB-231, (B) Hs578T, and (C) Hcc1143 cells than in 2D at 24 hours. BRD2 and BRD3 expression was not significantly changed for MDA-MB-231 or Hs578T cells, but they were significantly higher for Hcc1143 cells in 3D cultures.
Brd3 and Brd4 were induced in 3D cultures
BET Proteins Brd3 and Brd4 were expressed in all three TNBC cell lines (Figure 2A). Quantification of band intensity determined that MDA-MB-231 had 80% increase in Brd3 expression in 3D culture and Hcc1143 had three times expressions of Brd3 and Brd4 at 3D culture compared to 2D (figure 2B). Hs578T did not have significantly different Brd3 and Brd4 expression when they were cultured under the two conditions (Figure 2B).
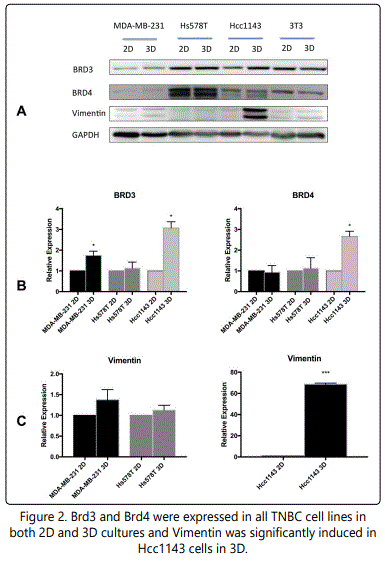
A. Western blots show that Brd3 and Brd4 were expressed by TNBC cells in both 2D and 3D cultures at 48 hours. 3T3 cells were used as a positive control. Significantly higher expression of Vimentin by Hcc1143 cells in 3D cultures suggests the presence of EMT.
B. Quantitation of the Western blot data shows increased Brd3 expression in MDA-MB-231 and Hcc1143 cells in 3D cultures and increased Brd4 expression in Hcc1143 cells in 3D cultures relative to 2D cultures.
C. Vimentin expression was significantly increased in Hcc1143 in 3D cultures.
EMT was induced in Hcc1143 cells in 3D cultures
To detect the presence of epithelial-to-mesenchymal transition (EMT) Vimentin protein expression was examined. After 48 hours the basal-like Hcc1143 cells had 70 times higher vimentin expression in 3D culture than 2D cultures (Figure 2A, Figure 2C). This suggested that EMT occurred Hcc1143 cells when they were cultured in a 3D culture. However, there wasn't a significant change in vimentin expression for the mesenchymal-like MDA-MB-231 and Hs578T cell lines when they were cultured in 3D.
JQ1 inhibited the growth of TNBCs
JQ1 is a selective BET bromodomain (BRD) inhibitor that inhibits the expression of Brd3 and Brd4. MTT assays were performed to verify the JQ1 inhibition of TNBCs using carboplatin as positive control. Figures 3 and 4 show the cell viability of TNBCs with JQ1 and carboplatin treatment, respectively, in 2D. The IC50 of carboplatin for MDA-MB-231, Hs578T and Hcc1143 cells were 18.2 μM, 5.3 μM and 10.8 μM, respectively (Figure 3). This study found that the IC50 of JQ-1 for MDA-MB-231, Hs578T and Hcc1143 cells were 37.4 ± 6.7 nM, 113.5 ± 8.7 nM and 162.6 ± 12.6 nM, respectively in the 2D culture system (Figure 4). All three cell lines were more sensitive to JQ-1 than to carboplatin in 2D cultures.
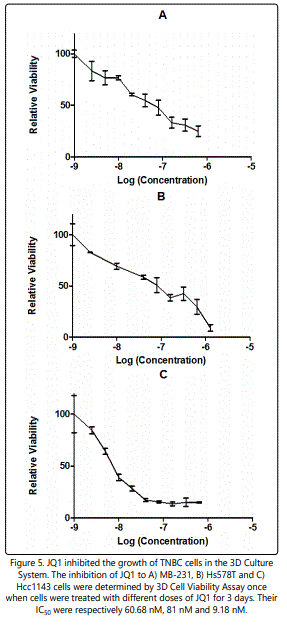
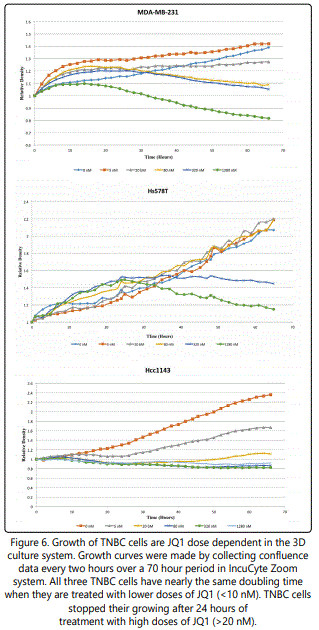
3D viability assays determined the IC50 of JQ1 for MDAMB-231, Hs578T and Hcc1143 cells as 60.68 nM, 81 nM and 9.18 nM in 3D cultures (Figure 5). Cell confluence in response to JQ1 treatment was assessed using an IncuCyte ZOOM™ Kinetic Imaging System. Figure 6 presents growth curves from the IncuCyte Zoom System showing the cell confluence of MDA-MB-231, Hs578T and Hcc1143 were all increasing slowly at lower doses of JQ1 (<20 nM) and dramatically dropped at the higher concentrations of JQ1 (> 20 nM).
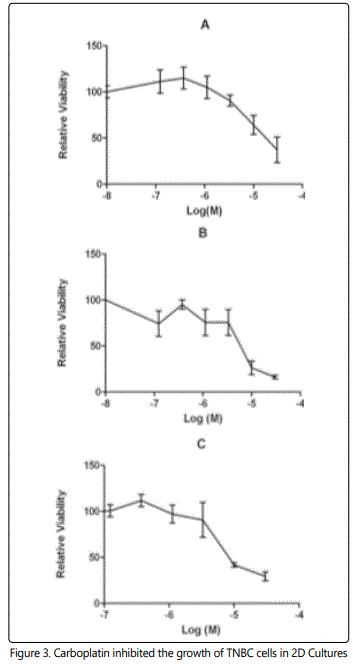
Carboplatin inhibition of A) MB-231, B) Hs578T, and C) Hcc1143 cells were determined by MTT Assay. Cells were treated with different doses of JQ1 for 3 days. Their IC50 were 18.2 μM, 5.3 μM and 10.8 μM, respectively.
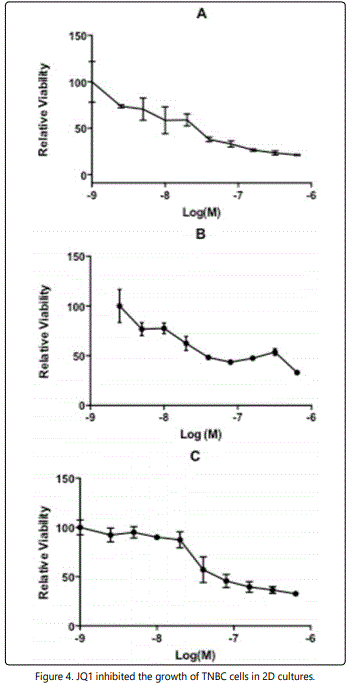
JQ1 inhibition of A) MB-231, B) Hs578T and C) Hcc1143 cells was determined by MTT assay. Cells were treated with different doses of JQ1 for 3 days. Their IC50 were 37.4 ± 6.7 nM, 113.5 ± 8.7 nM and162.6 ± 12.6 nM, respectively.
Discussion
Because of the lack of standard therapeutic targets in TNBCs, there is no effective receptor-based chemotherapy [2]. At present, the only drug approved by the FDA to treat TNBCs is carboplatin, which works by interfering with DNA repair [18]. However, the CALGB 40603 study (2014) concluded that additional treatment with carboplatin as the standard neo adjuvant chemotherapy after surgery for TNBCs increased the number of women who had a pathologically complete response [19]. Yet, the GeparSixto study (2014) acknowledged that the addition of carboplatin in the standard neo adjuvant chemotherapy for TNBCs improved disease-free survival from 76.1% to 85.5% [20]. Both studies unfortunately were not large enough to tell if adding carboplatin to the treatment of triple-negative breast cancer offered other benefits.
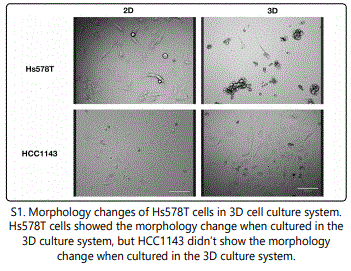
To model TNBCs, three different triple negative epithelial cells lines from breast carcinoma were used. MDA-MB-231 was derived from pleural effusion. Hs578T and Hcc1143 were derived from solid mammary gland tumors. The comparison of cells cultured in 2D and 3D systems showed differences in differentiation, gene expression, protein expression, and cell viability [21]. When Hs578T cells were cultured in the 3D culture system, the cells formed spheroid-like structures and clusters (S1). For HCC1143 cells, there was no morphology change when they were cultured in the 3D culture system (S1). The morphological changes in TNBC indicated that that Hs578T were mesenchymal-like, while HCC1143 cells were more basal-like TNBC [22].
Gene expression of BRD2, BRD3 and BRD4 was measured by qPCR. We found that MDA-MB-231 and Hs578T had significantly higher BRD4 expression in 3D. Hcc1143, however, had significantly higher expression of all three genes in 3D. Protein expression of BRD3 and BRD4 was found in all three TNBC cell lines. Brd3 was found to have an 80% increase in MDA-MB-231 cells and a 200% increase in Hcc1143 cells in the 3D culture system. Brd4 was found to have a nearly 300% increase in Hcc1143 cells in the 3D culture system. All these increases make it probable that EMT or an increased migratory potential can induce Bromodomain expression in the 3D culture system, especially for basal-like cells.
The development of other cancers have been related to dysfunction of BET bromodomain proteins, and BRD inhibitors have shown efficacy in several cancer models [23]. JQ1 can displace BET bromodomain proteins, such as Brd4, from chromatin by competing with acetyl-lysine recognition modules leading to the inhibition of oncogenic transcriptional programs [24]. Indeed, JQ1 induced a dose-dependent decrease in cell viability. The inhibitory effect of JQ1 on TNBC cell viability had an IC50 from 50 nM to 120 nM in the 2D cultures. Compared to carboplatin, JQ1 is about 1000 times more effective, which suggests that JQ1 would a potential effective therapy for TNBC. The IC50 of JQ1 for TNBCs in the 3D cultures was about 50 nM, demonstrating the effectiveness of JQ1 in the 3D cultures. IC50 of JQ1 to Hcc1143 decreased from 160 nM in 2D to 9 nM in 3D, indicating that Hcc1143 is more sensitive to JQ1 in the 3D cultures. This is interesting as the Hcc1143 cells has the most extensive shift in vimentin expression when moved to 3D cultures. It is also possible that the presence of EMT in 3D cultures increases the sensitivity to JQ1. Vimentin, related to EMT in breast cancer, can therefore be used as a biomarker for this sensitivity.
JQ1 binds to acetylated lysines blocking BET protein binding, but the function of JQ1 lacks selectivity within the BET family. This makes it difficult to clarify contributions of each protein family member to transcriptional and cellular outcomes [25]. The detailed pathways related to BRDs on TNBCs may still be unknown, but some promising findings resulted in an effect of JQ1 on the BRDs pathway and the relationship among Brd2, Brd3 and Brd4.
Hnilicová (2013) reported that the C-terminal domains of Brd2 were associated with chromatin modifying gene splicing, and Brd4 stimulated constitutive splicing of primary response genes likely via modulating RNA polymerase II C-terminus phosphorylation [26]. The knockdown of Brd2 was shown to result in reduced expression of almost half of alternative splicing targets because Brd2 influenced the formation of the initiation complex at the promoter [26]. The genes of the promoter stimulated transcription and regulated alternative splicing at the same time. However, the molecular mechanism of Brd2 mediated splicing regulation is also unknown.
It is also worth mentioning that expression of cell cycle regulators p21 and p53 have been found related to BRD2 expression. p21 transcription decreased ~60% and p53 increased ~60% after the knockdown of BRD2 [26]. It's likely that JQ1 inhibits the expression of Brd2 which then effects the transcription of at least 1400 genes, causing the termination of cell communication, cell differentiation, regulation of cell adhesion and macromolecule modification [26]. JQ1 was found to inhibit the expression of Brd4, a dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix [23]. Brd4 binds to the c-MYC promoter, an action that is reversed by JQ1.
Studies have shown that Brd4 inhibition leads to cell cycle arrest, senescence, p21 upregulation and apoptosis, all processes mediated by p53, a requirement for c-MYCdependent cell cycle arrest and differentiation, but not apoptosis [27-30]. Still, JQ1 does not exhibit a direct effect of Brd4 inhibition on p53 protein levels [10,27,31]. A finding consistent with the actions of JQ1 where ectopic expression of c-MYC in hematological cell lines confers significant resistance to JQ1-induced cell cycle arrest and differentiation, but cell death is not affected [27]. It has also been determined that JQ1 cooperatively removes Brd4 from chromatin and increases re-assembly of the 7SK snRNP, which can silence the transcription of c-Myc, resulting in growth arrest and apoptosis of multiple myeloma cells [32]. Synergistic effects of histone deacetylases and JQ1 could possibly be exploited for better treatment of these conditions and for future interventions for TNBCs.
In the other research, it was also determined that after the knockdown of BRD2, the transcription of BRD4 declined by almost 75%, whereas BRD3 expression decreased by ~40% which suggest possible cross-regulation of BET proteins [26]. Most likely, the 3D cell culture model will have different effect on the expression of Brd2 in different cell lines. The coregulation among Brd2, Brd3 and Brd4 could result in different expression of BRDs in different cell lines. Brd2 and Brd3 associated chromatin is significantly enriched in H4K5, H4K12 and H3K14 acetylation and contains relatively little dimethylated H3K9 [33]. Several studies focused on BRD2 and BRD4, but little is known about the properties of Brd3. LeRoy et al. (2008), found that Brd2 and Brd3 bind actively transcribed, acetylated chromatin along the cyclin D1 gene [33,34]. They also determined that Brd2 and Brd3 facilitate RNA polymerase II transcription through acetylated nucleosomes [33,35].
Our study found that Brd3 and Brd4 transcripts are present and the proteins expressed in TNBC cells. In our 3D culture system, cells were in a mesenchymal state, which induced the expression of bromodomain proteins. With the higher expression of BRDs, TNBC cells were more sensitive to the JQ1. JQ1 can inhibit the growth of TNBCs, but the underlying pathway was not studied here. However, based on the other studies, we propose the potential pathway would be Brd2-p21/p53 pathway and Brd4-cMYC pathway [36,10,26,27]. It has been shown that MYC can induced EMT in breast cancer cells [30], and Hcc1143 cells in 3D cell culture system have been shown in the transition from epithelial to mesenchymal, which suggests that HCC1143 cells have higher MYC expression in 3D cell culture system. JQ1 is BET inhibition and targets MYC dependent in cancer cells [31]. The overexpression of MYC induces the expression of BRD4, and EMT also recruits BRD4 to direct WNT5A expression in basal-like breast cancer [32]. BRD4 induction caused by MYC and EMT make HCC1143 cells more sensitive to JQ1 in 3D cell culture system.
Conclusion
JQ1 inhibits the growth of TNBC cells in both 2D and 3D cell culture systems. Our data suggests that cells that are more mobile have an increased sensitivity to bromodomain inhibitors. This provides a possible target for early metastasis of triple negative breast cancers.
Acknowledgment
We extend our gratitude to Dr. Christina Zito and Dr. Eva Sapi for the great support and suggestions on this study, and we also thank Dr. Gil Mor for the use of the IncuCyte Zoom.
References