Review Article
Response to Epigenetic treatment in Gastric Cancer Cells: a Mini Review
The New Lahijan Scientific Foundation, Iran
*Corresponding author: Saeed Taheri, The New Lahijan Scientific Foundation, 13166-44158, Iran, E-mail: taherimd@gmail.com
Received: May 1, 2018 Accepted: May 22, 2018 Published: May 28, 2018
Citation: Taheri S. Response to Epigenetic treatment in Gastric Cancer Cells: a Mini Review. Madridge J Cancer Stud Res. 2018; 2(1): 40-46. doi: 10.18689/mjcsr-1000105
Copyright: © 2018 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Introduction: Gastric cancers (GC) are usually considered associated with poor survival and significant resistance to chemotherapy in either clinical or in vitro settings, and epigenetic factors have been shown to be able to predict sensitivity of GC cells to therapeutic regimens. In the current study, the aim was to review the available evidence of the feasibility of epigenetic treatment including DNA methylation inhibitors (DNMTi) or histone deacetylase inhibitors (HDACi) on survival indices of GC cells in vitro.
Methods: The literature has been searched to find studies assessing potential impact of epigenetic drugs including DNMTi and HDACi agents on the survival and proliferative indices of GC cells, in an in vitro setting using Medline/Pubmed search engine. Finally 16 studies have been found, and their data got extracted and reviewed.
Results: Both DNMTi and hDACi drugs have been shown to significantly repress either proliferation or apoptosis indices of GC cell lines compared to untreated controls. However, when comparing to specimens under chemotherapy, they represented less effectiveness. The highest effect was observed when a combination therapy using both chemotherapy and epigenetic treatment was employed.
Conclusion: Epigenetic treatment is shown to be feasible in the management of GC cells in vitro, especially when co-administered with chemotherapy agents. Future studies in the clinical setting are recommended for confirmation of the results.
Keywords: Epigenetic treatment, Gastric Cancer Cells, Chemotherapy, DNA methylation inhibitors, histone deacetylase inhibitors.
Introduction
Epigenetics is the terminology used for defining heritable molecular mechanisms altering gene expression patterns without affecting DNA sequences, and they include DNA methylation, histone modifications, regional nucleosome load, and non-coding RNAs [1]. Epigenetic regulation of gene expression is a major factor in maintaining health control in the cell physiology, and it has been demonstrated that its alterations are major contributors in provoking and establishing aberrant gene expression profiles prevalently observed in the cancer cells.
More recent evidence is suggestive that epigenetic factors can be good indicators for several survival indicators in cancer patients including resistance or sensitivity to chemotherapy [2]. Gastric cancers are usually diagnosed in the late stages and they are associated with poor survival and significant resistance to chemotherapy in the clinical and in vitro settings, and epigenetic factors have been shown to well predict sensitivity and resistance of gastric cancer cells to different therapeutic regimens [3, 4]. To address this issue and considering their potential impact on reactivating tumor suppressor genes (TSGs), epigenetic drugs such as DNA methylation inhibitors (DNMTi) or histone deacetylase inhibitors (HDACi), have been the principle of intensive research in cancer treatment, and in some cases, they have actually been approved for the clinical use [5]. In the current study, the aim was to review the available evidence of the feasibility of epigenetic treatment on gastric cancer cells, and also some attempt has been made to cumulate results from different studies to make a conclusion.
Methodology
Basic Strategy and Search Engines Employed
The literature has been searched to find studies assessing potential impact of epigenetic drugs including DNMTi and HDACi agents on the survival and proliferative indices of gastric cancer cells, in an in vitro setting. Medline/Pubmed search engine was used for the search and Google Scholar for further investigation of the citations. No time limit was set for the searches. The following combination of terms has been used for search: methylation + gastric cancer, methylation + gastric cancer + chemotherapy, methylation + gastric + sensitivity, acetylation + gastric cancer, acetylation + gastric cancer + chemotherapy. Then the search had been repeated for each of the main HDACi and DNMTi agents separately. The literature search was originally carried out from 11 to 24 August 2017 and then renewed once more from 1 to 4 October 2017. Overall 16 articles [7-22] were finally found eligible to be included in this review article. Definitions of the terminologies and methodology in more detail have been previously described [6].
Inclusion and Exclusion Criteria
Inclusion criteria were: (1) To examine significance of either DNMTi or HDACi therapy in GC cell lines; (2) The studies' setting should have been in vitro; (3) To study either cell survival/proliferation indices or apoptosis in the study, and (4) To compare the results with controls or chemotherapy. Exclusion criteria were: (1) The study doesn't include data from in vitro setting, (2) The cells used are not from GC lines or data from the GC cell lines could not be retrieved from the cumulative data, and (3) If the study was not an original report (including review articles, meta-analyses, editorials, or so).
Accumulation of Data & Meta-Analysis
To the best of the author's knowledge, in studies as this one, Meta-analysis has never been conducted in the published literature, and one may think it is not a feasible analysis in this setting. Nevertheless, making some presumptions in the current study and despite the limitations, the author tried to conduct meta-analyses separately for either continuous data (attributed to mean±SD cell viability, growth, and survival percentages) or apoptosis in which percentages of apoptotic versus non-apoptotic cells have been used to conduct meta-analysis. To add a result row in the analyses, for each cell type and/or chemotherapy agent type, a new row has been added to the tables and the report got included into the final meta-analysis, and since each study was using different experiments (i.e. cell types, comparison chemotherapy drugs, but not for different dosages) to study potential impact of epigenetic drugs on GC cell activity indices, each study report could participate more than once in each meta-analysis (tables). Since different reports followed more or less consistent approaches in their methodologies, the author took the liberty to conduct meta-analysis employing percentages of apoptotic cells (versus actual number of the cases in each group which was not feasible here) compared to the non-apoptotic cell percentages. In this case, when the cell percentages included decimals, the nearest integer for each of them was used to be included in the meta-analysis.
Meta-analyses attributable to the continuous factors (including cell viability, growth, and cell survival) might have been associated with even more controversy, so their meta-analysis data has been presented only in the Supplementary file 1). In these analyses, the number of contributors in both comparison groups was equally considered 6, representative of the number of well micro culture plates consistently used in these reports. Although it may be a matter of controversy, but one could argue that if nothing else, it censors the differential effects of the population volume in the study groups.
Since Relative risk returned by the analyses were not significantly affected by the number of contributors (if considered equal anyway), the relative risk had been reported in the respective tables. In the figures, the weight of contribution of each study were appeared because they were only minimally affected by the number of each group contribution, while standard deviations were censored from the figures, because of its significant dependence on the contribution volume of the groups.
Result
DNA Methylation Inhibitors (DNMTi)
Extensive evidence indicates that DNA methylation in tumor suppressor genes (TSGs) is a major contributor to the altered gene expression profiles in cancer cells; therefore, pharmacologic inhibition of inappropriate gene methylation could be considered a potentially effective anti-malignancy approach due to its effect on the upregulation of gene expression at the TSGs.
DNMTi vs. Untreated Control
Some of the reviewed studies investigated potential impact of DNMTi alone or in combination with chemotherapeutic agents on GC cell growth, viability and apoptosis rates. Among the experiments comparing effects of DNMTi versus untreated controls, all but one, represented lower cell growth and/or viability rates for the specimens under DNMTi therapy than control specimens, though not all of them reached significance level (table 1.a). The only experiment with a conflicting result was on MKN28 GC cell line which is considered sensitive to cisplatin therapy (table 1.a). Similar observation was made in experiments comparing apoptosis rates in specimens under treatment with DNMTi vs. untreated control, with MKN-74 cell lines showing a lower apoptosis rate among specimens under DNMTI (vs. controls; table 1.b).
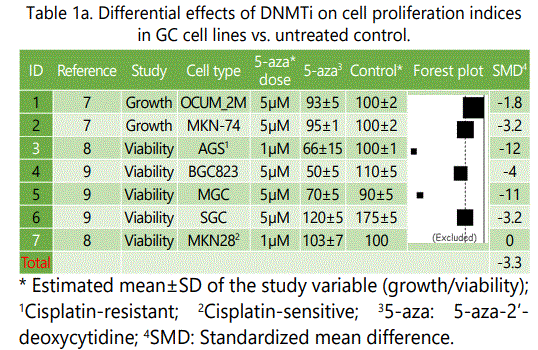
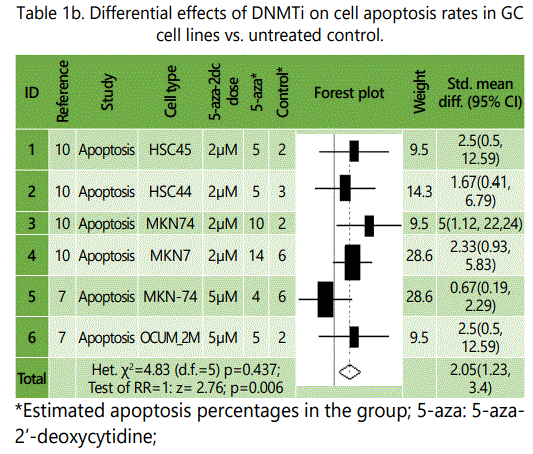
DNMTi Versus Chemotherapy
Although chemotherapy besides surgery is usually considered the backbone of patient management in the GC, however it is associated with broad spectrum side effects, and also it is not effective in a large number of patients. DNMTi monotherapy has been extensively investigated in the hematological malignancies, nevertheless, its efficacy in solid tumors has been a matter of controversy. Epigenetic drugs including DNMTi, despite their own side effects, have been shown to be safer in the cancer patients, thus if we find even comparable efficacy in their anti-cancer activity versus chemotherapy, they can be well replace it in the management of cancers. Table 2.a lists experiments comparing effects of DNMTi versus chemotherapy drugs on the cell proliferation indices of GC cell lines.
As it is shown in table 2.a, excepting two experiments (IDs, 1&2), in all the other experiments chemotherapy was associated with higher effectiveness than DNMTi therapy alone, and in the first experiment (Study ID: 1), dimethyl sulfoxide (DMSO) therapy might be considered not enough potent compared to the conventional chemotherapy agents studied in the other experiments. Although cisplatin represented no significant differential effect to DNMTi, oxaplatin, another platinum-based agent was associated with significantly higher effect. The largest differential effect was observed for gemcitabine versus DNMTi (table 2.a). Table 2.b summarizes differential apoptosis rates reported for GC celllines treated by either chemotherapy or DNMTi agents. As it is well shown in the plot, all the experiments showed significantly higher pro-apoptosis effects in favor of chemotherapy.
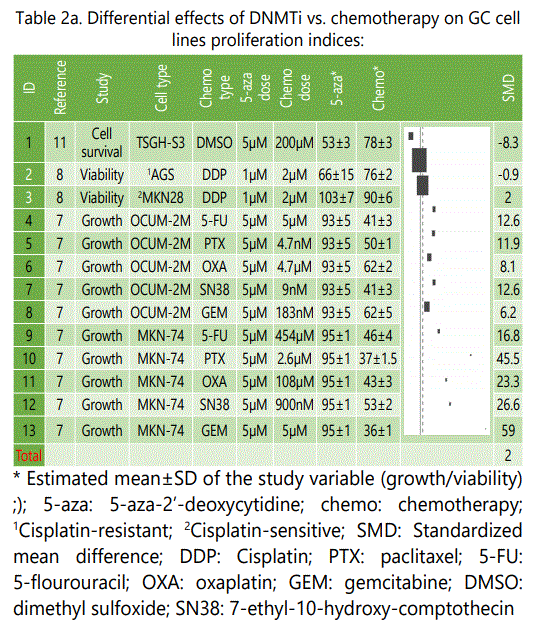
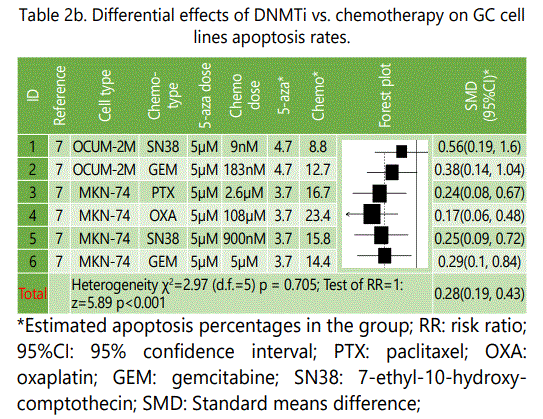
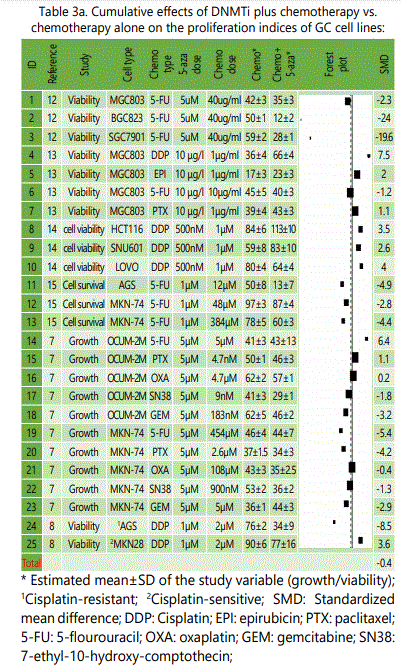
DNMTi Plus Chemotherapy Versus Chemotherapy Alone
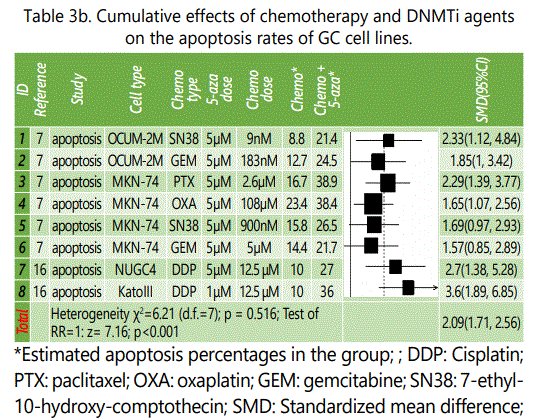
Table(s) 2 showed that chemotherapy is more efficacious in either repressing proliferation indices or apoptosis induction in GC cell lines than DNMTi agents. However, since they are likely to do in different pathways in inducing their anti-malignancy effects, a combination therapy using both chemotherapy agents as well as DNMTi agents may not only increase anti-cancer effects of therapy but also it may provide the opportunity to decrease dosages of either drugs and attenuate their potential side effects. Moreover, these combinations might promise synergic effects outweighing their cumulative effects alone. Table 3.a summarizes data of experiments comparing cumulative effects of chemotherapy and DNMTi versus chemotherapy alone in GC cell lines. As it is shown in the plot, experiments employing 5-FU plus DNMTI (versus those under chemotherapy alone) seem to be showing higher anti-proliferation efficacy compared to the combination therapy with platinum based chemotherapy, indicating that combination therapy of GC cell lines using DNMTi agents and 5-FU might represent more additive/synergistic effects on the prohibition of proliferation in these cells than platinum based regimens. It should be considered that this finding is not based on direct statistical analyses which is not possible in this study, but is based on the difference observed in the plot; therefore, making any scientific conclusion on it needs confirmation by direct comparison in future studies. Table 3.b lists results of experiments comparing apoptosis rates in the two study groups and shows in all the experiments, combination therapy using DNMTi and chemotherapy is significantly more efficacious in inducing apoptosis in the GC cell lines.
Histone Deacetylase Inhibitors (hDACi)
Histone deacetylases (hDAC) are enzymes that catalyze the removal of acetyl groups from the lysine residues from histones and there are 18 hDAC enzymes functioning in the human body. Histone acetylation has been shown to be an important epigenetic factor inducing gene activation, and therefore theoretically, as like the DNMTi drugs, inhibitors of hDAC enzymes can be used to reactivate TSGs repressed in cancer cells. In the clinical setting, two hDACi drugs, vorinostat and romidepsin, have been approved by FDA for use against refractory T cell lymphomas, and many others are under investigation [17], however to date, no study has investigated potential effects of hDACi on the GC patients in clinical setting.
hDACi vs. Untreated Control
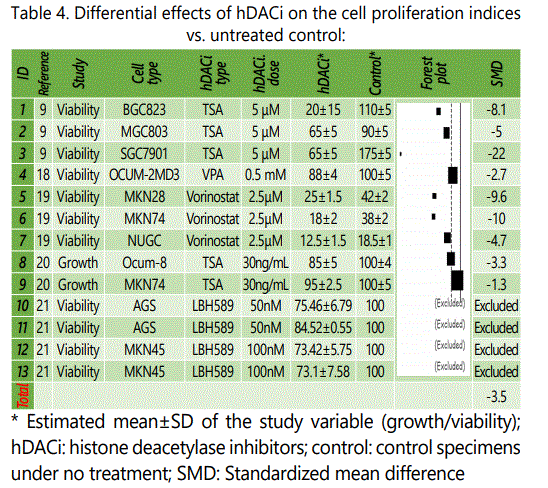
Table 4 summarizes data of experiments comparing effects of hDACi versus untreated control on the proliferation indices of GC cell lines. Data from 14 experiments from 5 studies have been presented [reff]. In all the experiments, mean ± standard deviation of the specimens under hDACi therapy were lower than their untreated control counterparts. Four experiments have been excluded from the analysis, because standard deviation for their mean cell viability rates of the control specimens had not been presented by the original report [21].
hDACi Versus Chemotherapy
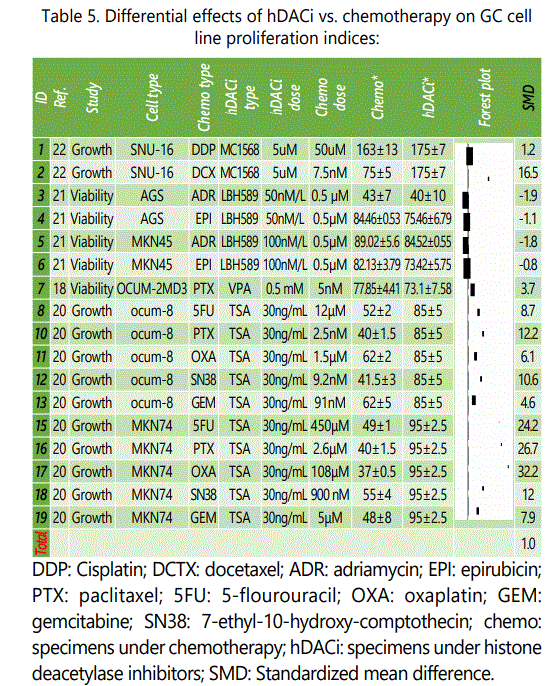
Anti-cancer effects of hDACi agents have also been investigated against chemotherapy in the GC cell lines (table 5). Unlike DNMTi, GC specimens under hDACi therapy, at least in some of the experiments, showed some minimally higher effectiveness versus chemotherapy alone, although at the end, just like DNMTi treated specimens, chemotherapy showed a superior effectiveness than hDACi (table 5). Whether it was related to the specific GC cell lines used in those experiments (AGS & MKN45) or the hDACi agents employed (LBH589) or the chemotherapy type needs further evaluations. These puzzle pieces are exactly what we need to find specific indicators of response to the epigenetic therapy, in order to maximize treatment effects in individual cancer patients.
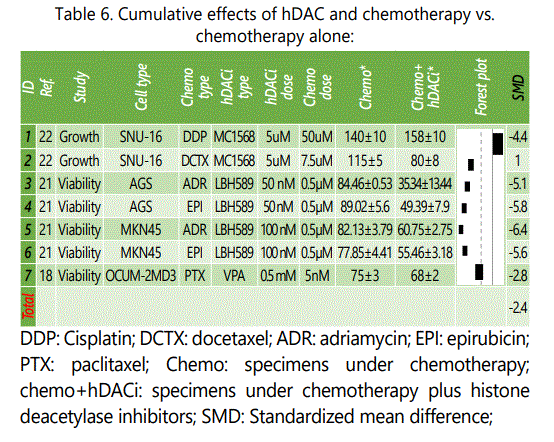
hDACi Plus Chemotherapy Versus Chemotherapy Alone
Three studies compared cumulative effects of chemotherapy and hDACi versus chemotherapy alone on the proliferation indices of GC cell lines in 7 experiments (table 6). Except in one experiment, in all the others, the cumulative effect was more powerful than treatment with chemotherapy (which was cisplatin in this case) alone. Exactly the same experiment employing another chemotherapy agent (docetaxel) showed showed a synergistic effect for the combination therapy. Apoptosis rates were also higher in specimens under hDACi plus chemotherapy versus chemotherapy alone. In an experiment on SNU-16 GC cell line, combination therapy with docetaxel and MC1568 represented mean±SD apoptosis rates of 6.6±2.2% versus 3.3±0.5% in specimens under docetaxel treatment alone [22]. In another experiment on AGS GC cell line, epirubicin plus LBH589 induced mean±SD apoptosis rate of 48±1.5% versus 23.3±2 for epirubicin alone [21].
Conclusion
In a previous study at the same issue of this journal, microRNAs, as an epigenetic factor, have been associated with response to therapy in the GC cell lines [23]. Such evidence promise that, alterations in the epigenetic profiles of cancer cells may also in turn be of therapeutic value in the management of cancer cell lines. To address this issue, the literature had been screened in this article, for reports investigating in vitro evidence for potential altering effects of epigenetic treatment of GC cell lines on their proliferation and survival indices. And finally it had been attempted to cumulate their data to provide a conclusion.
According to the findings of this study, epigenetic therapy of GC cell lines is associated with improvement in the GC cells' survival indices, although this effect was not comparable to that of chemotherapy. The best response in either DNMTi or hDACi drug therapy had been achieved when they were co-administered with chemotherapeutic agents, and the latter was more effective in downregulating GC cell proliferation and inducing apoptosis than either epigenetic treatment or chemotherapy, alone. A simple look at the forest plots, one can find exceptions to this conclusion. Whether these exceptions are related to the GC cell lines used, the chemotherapy drug or the epigenetic agent employed needs future evaluations. And finally before they can be considered of any practical use, all these findings need to be confirmed in the in vivo and clinical settings.
This study had been associated with some limitations. Although in this study, it has been tried to use the most possible orthodox methods for the analyses, nevertheless conducting Meta-analyses in studies of this nature is not conventional and will be a matter of controversy. However, considering the inconsistencies in the reports (i.e. see forest plot in table 3.a), one may think that there might be need for some type of Meta-analysis in the in vitro studies as wellin which the main playing factors could be the number of well microculture plates, the number of cells in each well, and how to use the percentages of cells respond or not to the intervention. Other confounding factors may include, how to include multiple experiments from the same study when it is performed on different cell types, different drug treatments, and especially when same experiments are repeated with different dosages of the same drug. Moreover, is it accurate to include experimetns that had been using the same control data as the comparison group (i.e. two experiments on two different chemotherapy drug treatments when their data is compared to the same untreated control). Another limitation of the analyses included in this study is that the time of treatment for each experiment was inconsistent between different studies, although similar treatment time length for the two wings of each experiment was consistent in all of them. Also, it should be considered that such inconsistencies exist in the conventional Meta-analyses as well. Despite the drawbacks mentioned for this study, some positive aspects can also be noted for it. Inclusion of reports from different GC cell lines representing differential response to epigenetic treatment could be a better representative of how GC patients generally respond to the same epigenetic drug therapy than individual studies, at least until a routine definition of the specific GC cell type for each patient is feasible, prior to therapy initiation. And finally all the results should be properly confirmed in the clinical setting before any definitive conclusion about the clinical values of its findings could be made.
References