Review Article
In vitro Evidence on Associations between MicroRNAs and Response to Therapy in Gastric Cancer: Report from the Encyclopedia Amlashica Systematic Reviews
The New Lahijan Scientific Foundation, Tehran, Iran
*Corresponding author: Saeed Taheri, The New Lahijan Scientific Foundation, Kordestan Str Tehran, Iran, Tel: 14376-45685, E-mail: taherimd@gmail.com
Received: September 25, 2017 Accepted: November 02, 2017 Published: November 08, 2017
Citation: Taheri S. In vitro Evidence on Associations between MicroRNAs and Response to Therapy in Gastric Cancer: Report from the Encyclopedia Amlashica Systematic Reviews. Madridge J Cancer Stud Res. 2017; 1(1): 12-33. doi: 10.18689/mjcsr-1000103
Copyright: © 2017 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Introduction: Gastric cancer (GC) is a malignant disorder with one of the highest lethality rates of all. Intensive efforts have been made to develop directed therapy approaches to individualized treatment to improve the outcome, yet not very successful. This systematic review aims to make a thorough understanding of the existing data in this way.
Method: The literature had been reviewed for all the in vitro evidence on potential associations between miRNA expression and response to therapy in GC. Finally, 108 studies reporting their experiment with 178 miRNAs were found and their data extracted. Reports of miRNAseffects on response to chemotherapy have been re-categorized into 4 subgroups: (1) effects on cell growth; (2) differential expression in resistant vs. parental cells; (3) response to elevating doses of drugs; and (4) apoptosis induction.
Results: miR-223, miR-196, miR-92 and miR-181a show the highest up-regulation rates in GC cell lines resistant to cisplatin vs. parental cells.The miRNAs with the highest apoptosis induction rates due to cisplatin therapy included miR-200b, -497, -200c, and -429 respectively, all have a common target of proto-oncogene Bcl-2. Allthe three miRNAs inducing the highest resistance to adriamycin in GC cell lines (-19a, -19b, -106a) have a common target of PTEN, while all the three antagomiRs (-200c, -218, -497) conferring highest sensitivity to doxorubicin target Bcl-2.Among miRNAs with the lowest IC50 modulation for agomiRs (miR-429 and -27b), both of them target Bcl-2; and the highest IC50 was found for antagomiRs antagomiR-27b & -23b-3p.
Conclusion: Our knowledge on the associates of GC treatment is very limited to make a safe conclusion of what drug to use in different situations, and therefore, future studies should mention this issue.
Keywords: In vitro, Micro RNAs, Gastric Cancer, Chemotherapy, PTEN
Introduction
Gastric cancer (GC) is globally the second leading cause of cancer-related mortality, and the blames are mostly directed at its insidious nature, which results in late stage diagnosis of the disease with poor therapeutic efficacy. Therefore, determining factors associated with resistance to therapeutic regimens as well as those that confer higher sensitivity is of extreme relevance.
Besides surgery which had been widely employed in the management of GC, systemic chemotherapy is a cornerstone of the treatment in these patients in either locally-invasive or metastatic disease, worldwide [1, 2]. Radiotherapy has also been occasionally reported as a useful procedure in these patients, especially in combination with chemotherapy. Nevertheless, despite the use of different potent therapeutic regimens developed for GC patients with all the promising outcomes reported by the clinical trials, the mortality rates are still very high and therefore, efforts have been made to invent individualized therapeutic strategies that target features of the disease in more distinctly defined GC subgroups [2, 3, 4].
MicroRNAs (miRNAs) are a class of non-coding RNAs of 19–25 (~22) nucleotides that regulate post-transcriptional processes of target genes through inhibiting translation of the target mRNAs by binding to their 3'untranslated region (UTR). MicroRNAs impose a substantial control on cell homeostasis, and their dysregulation has been associated with a broad spectrum of pathological processes including cell proliferation, invasion, metastasis, and apoptosis [4]. In studies on GC similar to several other malignancies, microRNA expression profiling has been extensively used in cell lines, tissue samples and serum in order to reveal the biological processes as well as clinical consequences in which microRNAs play significant roles [5]. A very large number of differentially expressed miRNAs have been revealed through these studies with a broad number of suggested biological functions, while the number of them conferring clinical use was limited. Tumor behavior including local invasion, lymph node dissemination, distant metastasis, and prediction of response to chemotherapy as well as providing prospects of individualized therapeutic landscapes are some of the clinical advantages that come from these studies. Encyclopedia Amlashica is an attempt to systematic reviewing the existing data on the genetic and epigenetic associations of response to therapy in cancers. The aim of the current study was to conduct a comprehensive and thorough review of the literature indicative of any potential role for microRNAs in the treatment of GC. As it can be conferred from the title, the author's intention was to make it an encyclopedia in this field, indispensible for all those who either work or intend to conduct prospective researches in this context.
Methodology
Basic strategy and search engines employed
A systematic literature search was conducted to ascertain studies assessing potential associations between microRNAs with resistance and/or sensitivity to therapeutic approaches in GC. MEDLINE was the premier database & search engine that was used for this purpose and in cases there was no link to the fulltext of an article in MEDLINE, Google Scholar was used for search; and for some individual papers, Google search engine or the journal websites have been attended. Due to the novel nature of our topic, no time limit was set for the searches (for none of the found studies, the publication date was before 2000; just one study before 2005). The literature search was carried out from 11 to 24 August 2017. To reinforce the power of research, citations to the most relevant articles found by MEDLINE search have been screened for more studies, potentially missed by the original method of search.
Search Terminologies
The following combination of terms have been used for search (terminologies used; total titles returned by search): (gastric + microrna + adriamycin; 14), (gastric + microrna + doxorubicin; 11), (gastric + microrna + cisplatin; 57), (gastric + microrna + fluorouracil; 38), (gastric + microrna + vincristine; 11), (gastric + cancer + microrna + resistance; 123), (gastric cancer mirna sensitivity; 163), (gastric cancer mirna radioresistance; 2), (gastric cancer mirna radiotherapy sensitivity; 4), (gastric cancer mirna radiation resistance; 5), (gastric cancer mirna radiation sensitivity; 6), and finally a thorough screening was performed by a more comprehensive search term of (Gastric + cancer + microRNAs; 1590).
Study Selection and Review Process
To be eligible, studies had to meet the following criteria: (1) to examine significance of chemotherapy or radiotherapy in GC; (2) provide data on microRNA expression in GC setting; (3) to report analysis of potential associations between miRNA expression and chemo-sensitivity; and (4) the study environment must have been in vitro; yet, data of studies also including in vivo or clinical data besides the in vitro setting were also reported. Studies were excluded if (1) data of gastric adenocarcinoma could not be retrieved from the results; (2) the study did not include in vitro setting; (3) the study did not include anything about "chemo-response" in GC cell lines; (4) in case of comparisons of miRNA expression levels in cell lines, the comparison was between tumoral and normal cells (vs. –resistant & -sensitive both tumoral cells); and (5) review articles, metaanalyses, editorials/letters, and case reports. Studies prepared as an abstract or congress proceeding have not been exluded. All the study titles, abstracts and full-text articles found by the electronic search were reviewed by the author. Reference lists from review papers and relevant articles were also examined for additional studies that met the inclusion criteria.
Data extraction
A systematic approach to data extraction was used to produce a descriptive summary of the study GC cell lines, interventions and findings. A checklist had been developed by the author and then data got extracted from each study. In case data were not available in numerals, estimation was made from the charts & figures, wherever possible. The extracted data had been rechecked by the author to detect any inconsistencies and potential mistakes in data extraction or entering.
Categorization of the definition methods for chemo-response
Because the reviewed studies had employed various methods for defining potential chemo-responsive effects of the investigated epigenetic factors, therefore, to make a more distinctive & organized report of this systematic review, a recategorization of the methods used for detecting chemoresponse in the reviewed studies has been invented (see below):
Method A: Differential response to a chemotherapeutic agent detected by alterations in the proliferation/growth rate or colony formation by either up- or down-regulation (i.e. by transfection) of the respective miRNA in a cell line;
Method B: Detection of the differential expression of any miRNA in a chemo-resistant vs. parental cell line;
Method C: Detection of changes in the trend of cell viabilityinduced by variations in the concentration of a chemotherapeutic treatment, and/or detection of IC50 (see below);
Method D: Detection of apoptosis rate induced by altered miRNA expression in the context of chemotherapy (see below).
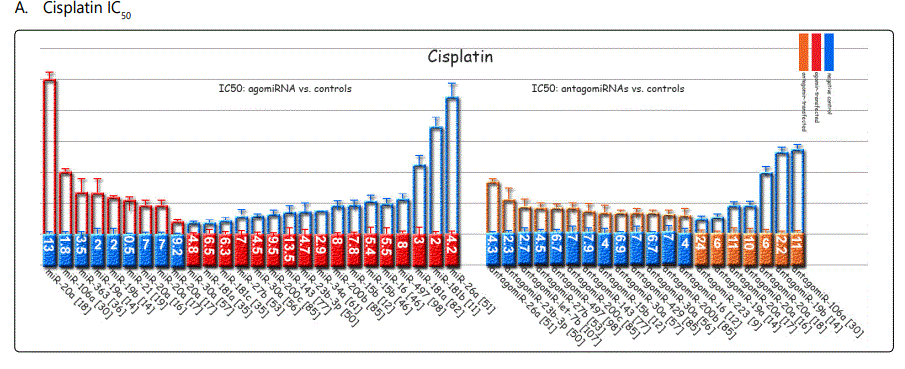
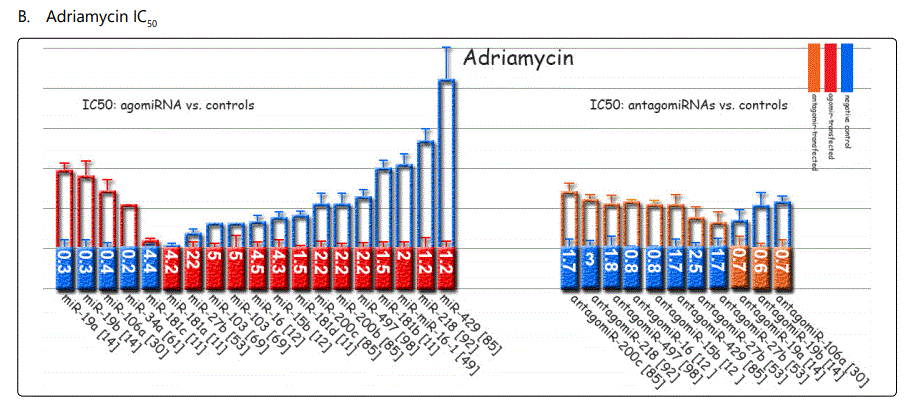
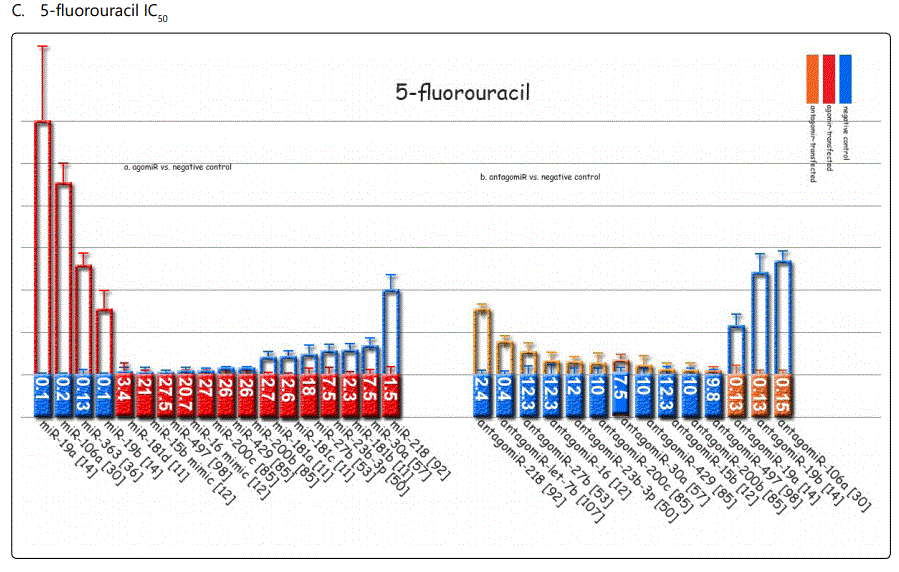
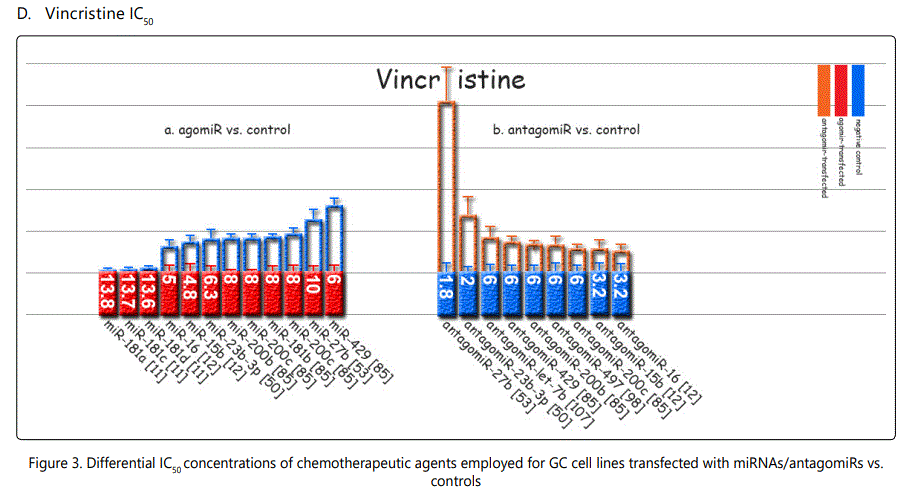
IC50
IC50 is an indicator of drug concentration at which cell growth is inhibited by 50%. So, each miRNA in any study can have distinct IC50 values for every chemotherapy drug. Figure 3 shows a schematic view of the studies providing detailed data on the differential IC50 values in GC cells transfected with either miRNAs or antagomiRs versus control negatives. Data presentation in this figure is not based on the actual values of IC50 in the study specimens, but is based on the fold difference in the measured IC50 for the two groups; therefore it is possible to anyone to have an overall estimation and also compare the magnitudes of each miRNA or antagomiR in inducing IC50 dysregulation in GC cell lines. In this regard, the comparison groups, either the miRNA/antagomiR or the negative controls that represented the lower concentration severed as the unite IC50, and were presented as the base column in the figure; while the other comparison GC cell group that represented larger IC50 concentration, were showed as the larger column, with the column length representative of the fold difference to the other group. So, it should be re-emphasized that the length of the columns in figure 3 are not representatives of the actual IC50 values, but they represent the fold difference in IC50 concentration values of cells transfected with either the miRNA/antagomiR or control transfected cells. The number values presented in the base column is an estimate of the actual IC50 concentration of the group representing the basic (lower) IC50 value. Thus, based on the length of the column above, one can simply calculate an estimation of the IC50 concentration of the second group as well. Because it was not possible to provide detailed data for all, in case readers doubt about the unit of the value appeared in the baseline column, they need to go to the original paper. For either IC50 or apoptosis reports, whenever there were experiments in chemo-resistant cells and parental lines, the parental lines have been selected for the report. Also, if similar study from several cell lines has been reported, the more conventional lines (i.e. SGC-7901, MGC803 or BGC823) were selected for reporting.
Apoptosis
Definition: Apoptosis can be measured by several methods, but in this systematic review, only apoptosis rates detected by most precise and conventional methods available were collected and reviewed: (1) Apoptosis rates determined by flow cytometry through staining cells with Annexin-Vfluorescein isothiocyanate (Annexin-V-FITC) apoptosis detection kits or the terminal nucleotidyl transferase-mediated nick end labeling assay (TUNEL) cell death detection kits.
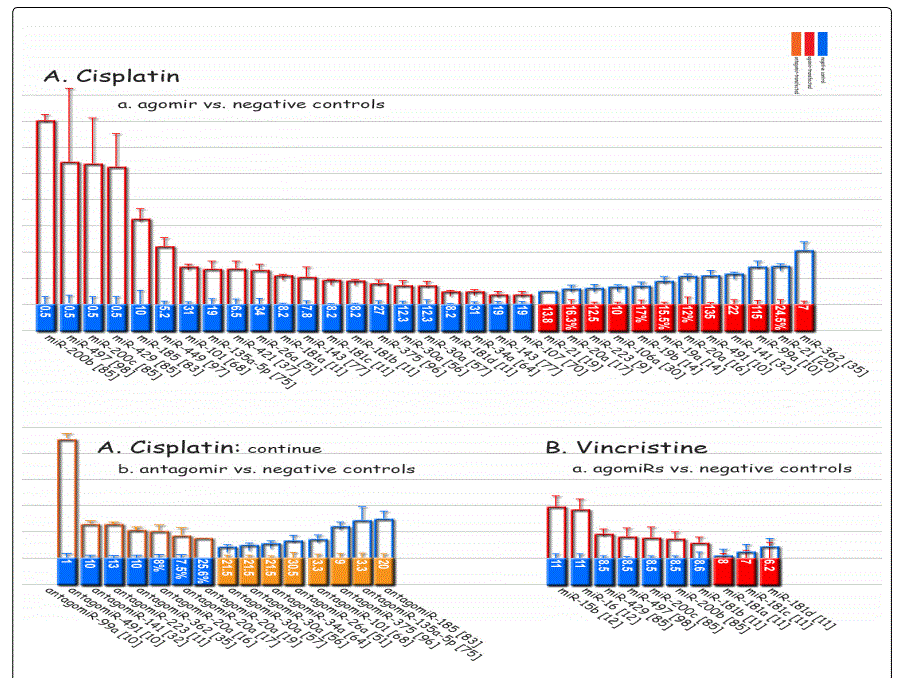
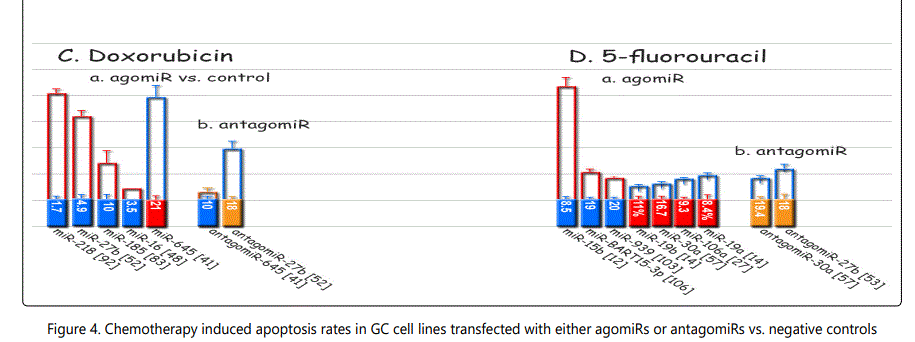
Data presentation: Figure 4 represents the differential apoptosis rates regarding miRNA expression rates in GC cell lines transfected with agomiRs/antagomiRs vs. negative controls. Again as mentioned in the 2.5.1. subsection for IC50, in this figure also, lengths of the columns do not represent the actual apoptosis rates of the GC cells, but only the fold difference in the estimated measured values for the respective GC cells versus controls. And also the values appeared in the basic column is an estimate of the actual apoptosis indices measurements.
In Vivo & Clinical Setting Studies
Some of the reviewed studies, besides there in vitro data, they also included in vivo setting. Nonetheless, the in vivo studies were mostly investigating issues not directly associated with the topic of the current systematic review; therefore, no detailed report on their results has been made in the current paper. Nonetheless, for those readers who are interested to get more data on the reports from their in vivo setting, all the studies have been screened for a potential in vivo setting report, and the found articles got marked in the tables. Some of the reviewed studies also included approaches reflecting clinical information on the functions of miRNAs in the anticancer drug resistance of GC patients; however, because it was out of the current study scope, and also the very large number of the studies on this setting, clinical data were excluded from this report, and has been reserved for a future report from the Encyclopedia Amlashica
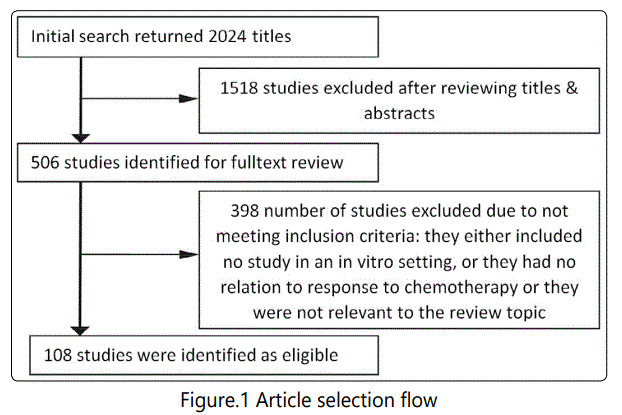
Results
Baseline Characteristics of Included Studies: A total of 2024 studies were identified primarily, from which 1518 were excluded after reviewing titles and abstracts. Then, 398 studies were excluded after full-text review. Finally, 108 studies were included (Figure 1) [6-113]. The included studies were all published between 2003 and 2017. Finally, 178 miRNAs with a report on their potential association with GC treatment efficacy in an in vitro setting have been identified. The miRNA association to chemotherapy response had been studied in the following methodologies:
(1) Studies solely investigating dysregulation of miRNAs in chemo-resistant vs. parental GC cell lines (method B):
(a) miRNA up-regulation: 41 unique miRNAs reported by 6 studies (table 1);
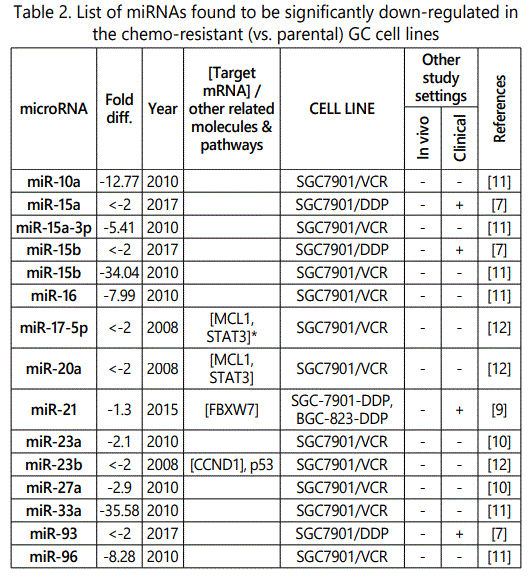
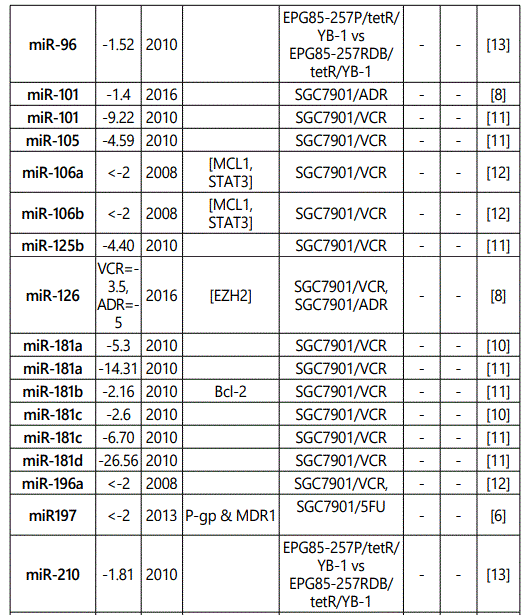
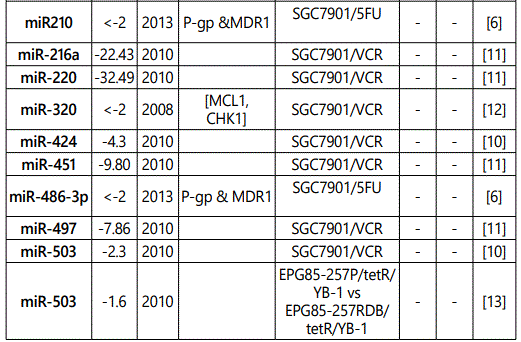
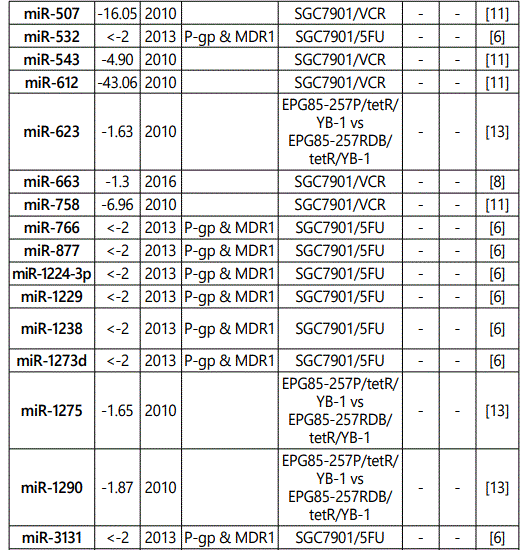
(b) miRNA down-regulation: 68 miRNAs (62 unique; 6 reported twice by two different studies: miR-101, miR-15b, miR-181a, miR-181c, miR-503, & miR-96) reported by 8 studies (table 2);
(2) Studies with reports on higher resistance to chemotherapy in cells with an miRNA upregulated expression or vice versa: overall 52 miRNAs (39; multiple reports: miR-27a (4 times), miR-223 (twice), miR-21 (6 times), miR-20a (thrice), miR-19b (twice), and miR-106a (twice)) from 33 studies have been reported in this category (table 3);
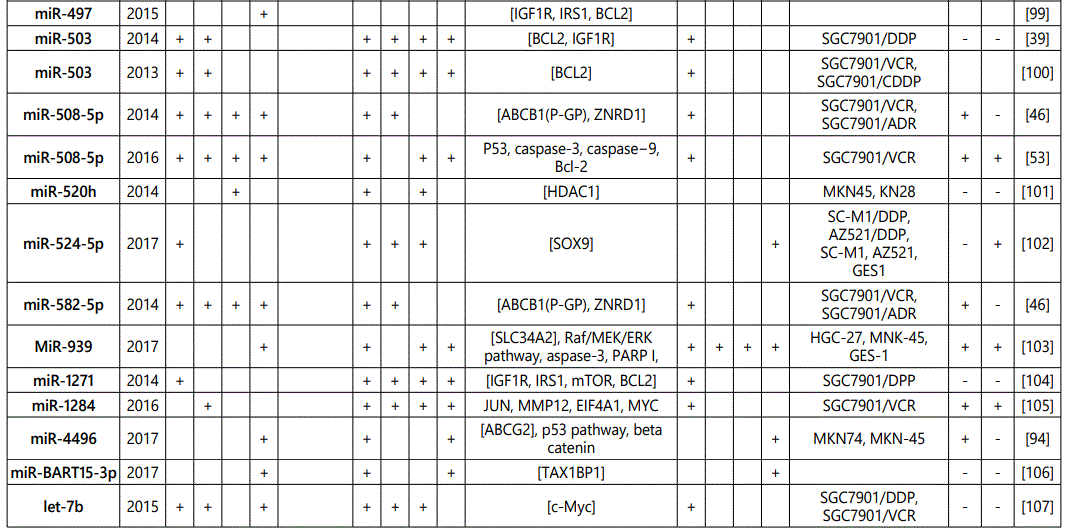
(3) Studies reporting higher sensitivity to chemotherapeutics in cells in which the miRNAin study was upregulated or vice versa: 92 miRNAs (69 unique miRNAs; miR-101 (twice), miR-107 (twice), miR-143 (twice), miR-15b (twice), miR-16 (4 times), miR-200b (twice), miR-200c (4 times), miR-204 (twice), miR-218 (twice), miR-27b (twice), miR-30a (twice), miR-34a (4 times), miR-429 (twice), miR-497 (twice), miR-503 (twice), miR-508-5p (twice), miR-7 (twice)) from 69 studies have been reported (table 4);
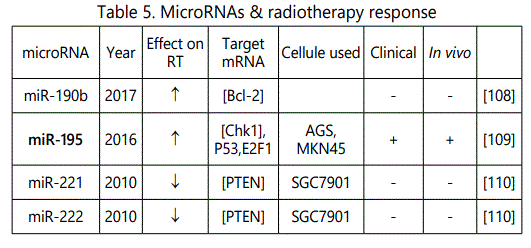
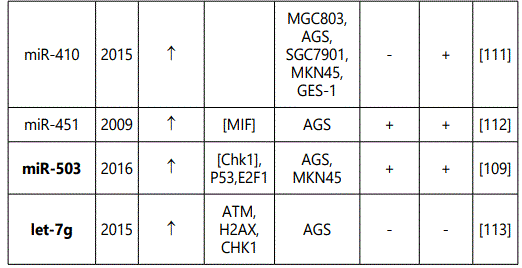
(4) Studies reporting alteration in resistance or sensitivity to radiotherapy in GC cell lines in which miRNA levels have been dysregulated: 8 unique miRNAs have been reported here by 6 studies (table 5);
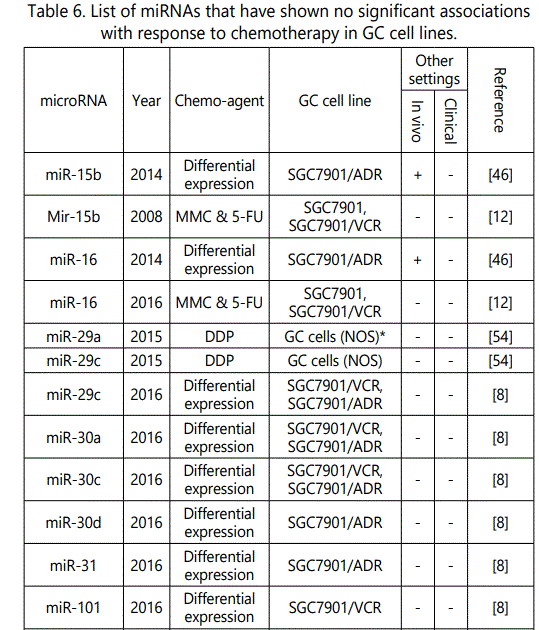
(5) Studies reporting miRNAs conferring no alteration in the response to chemotherapy in GC cell lines: 22 miRNAs (21 unique, miR-29c, Mir-15B & Mir-16, each reported by two studies) reported by 5 studies; (table 6).
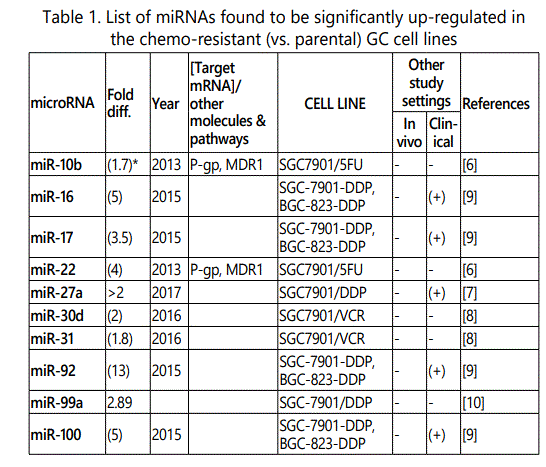
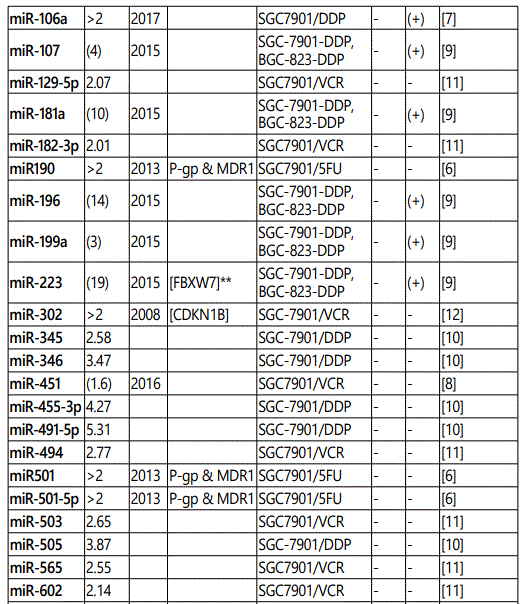
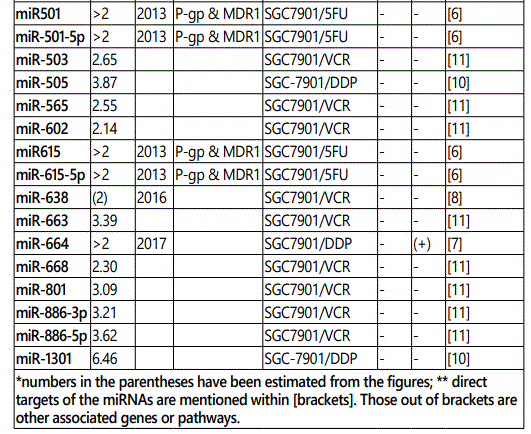
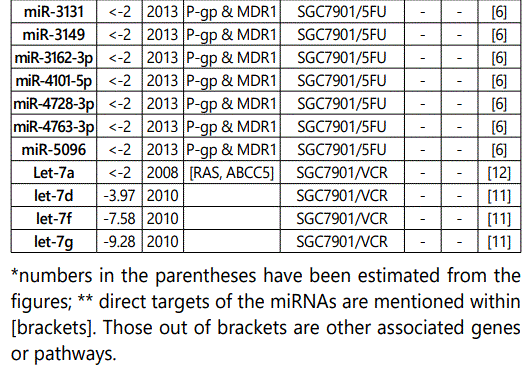
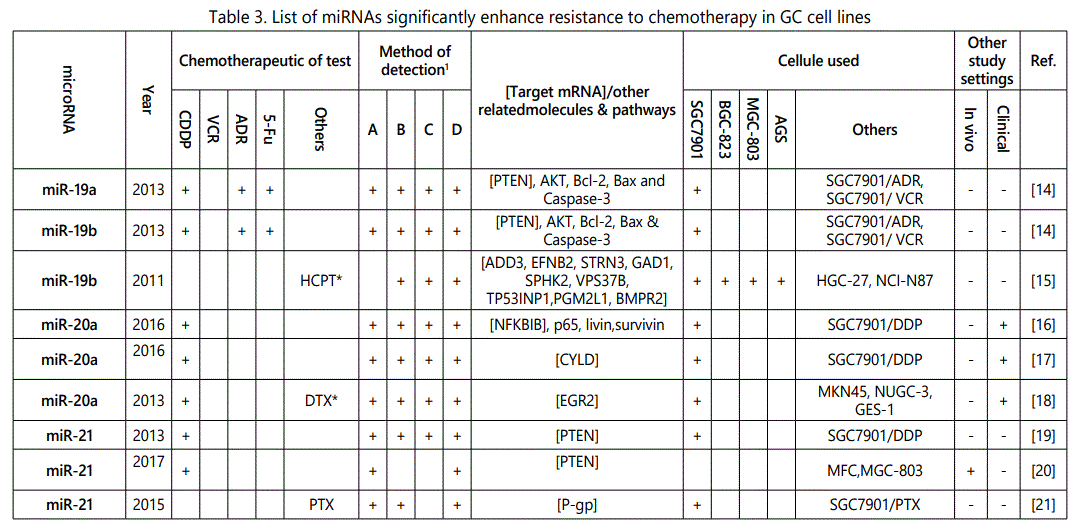
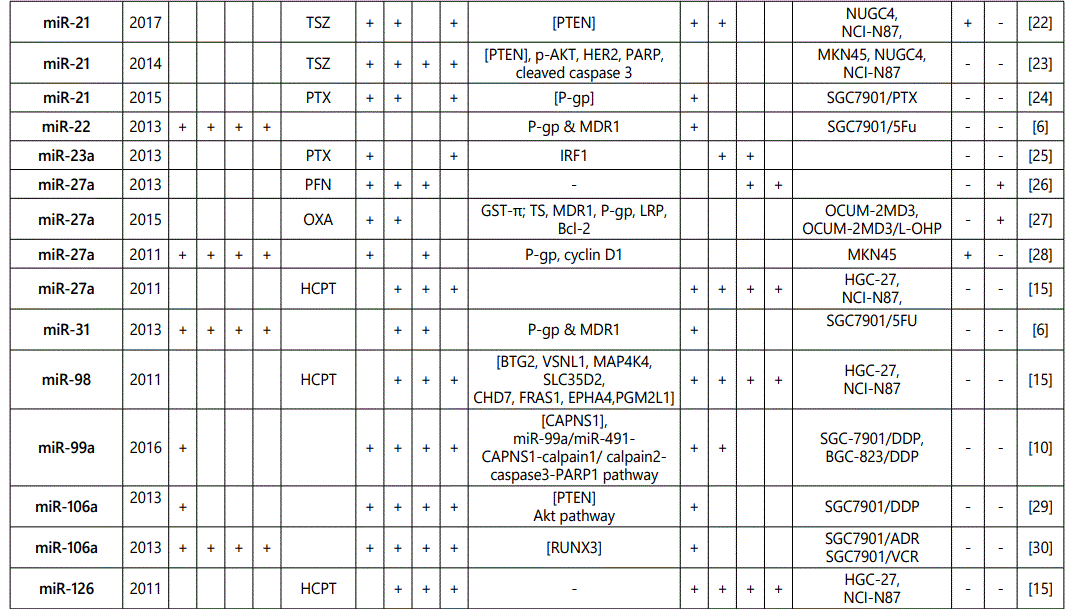
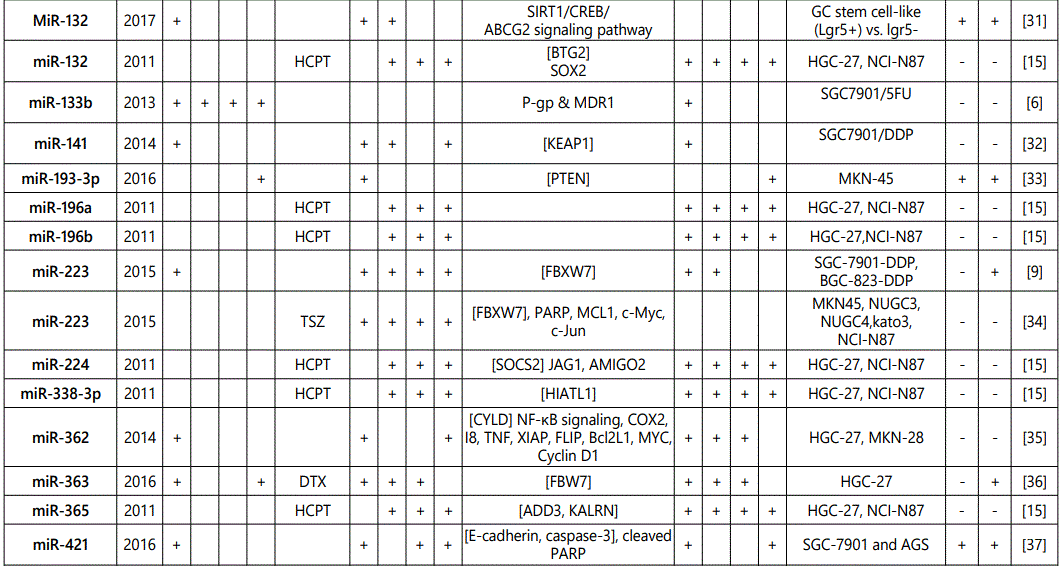
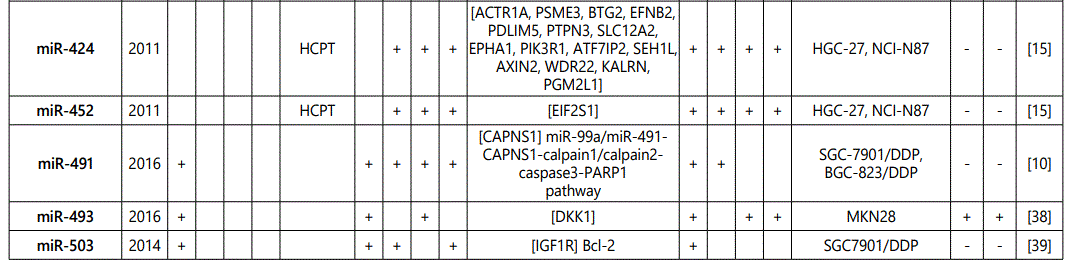
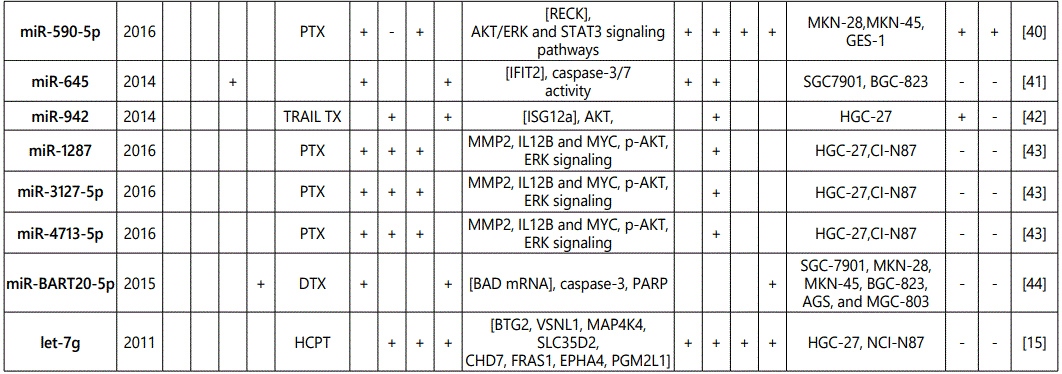
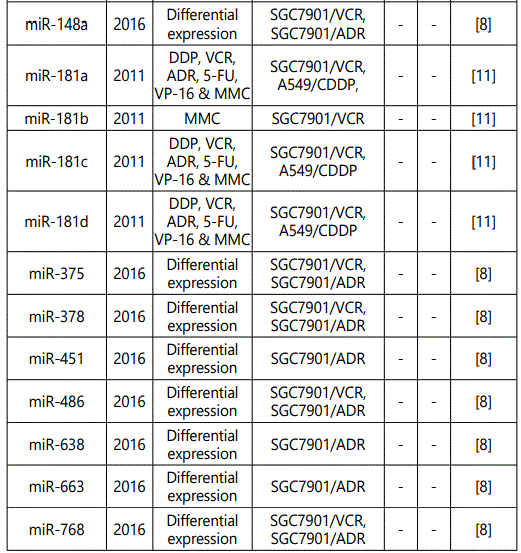
1Measurement methods for defining chemo-response (A-D): A: Growth rate/proliferation or colony formation; B: differential expression in resistant vs. parental GC cell lines; C: rate of growth inhibition by various drug concentratioor IC50 detection; D: apoptosis rate (detailed information can be found in the methods section).*HCPT: Hydroxylcamptothecin; PFN: Perifosine; DTX:Docetaxel; TSZ: trastuzumab; PTX: Paclitaxel; OXA: Oxaliplatin. Direct targets of the miRNAs are mentioned within [brackets]. Those out of brackets are other associated genes or pathways.
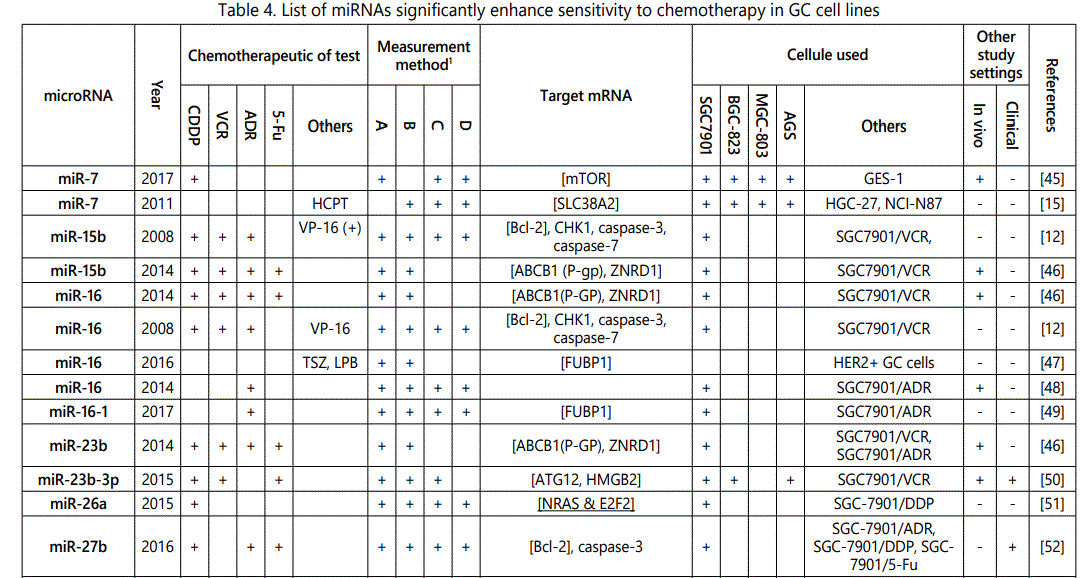
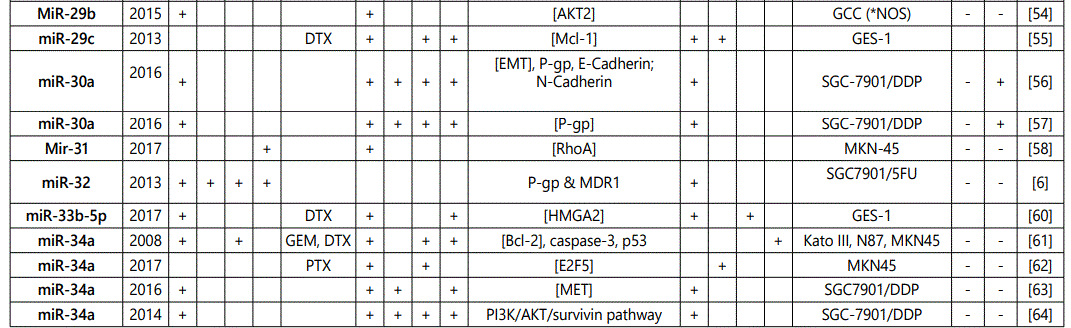
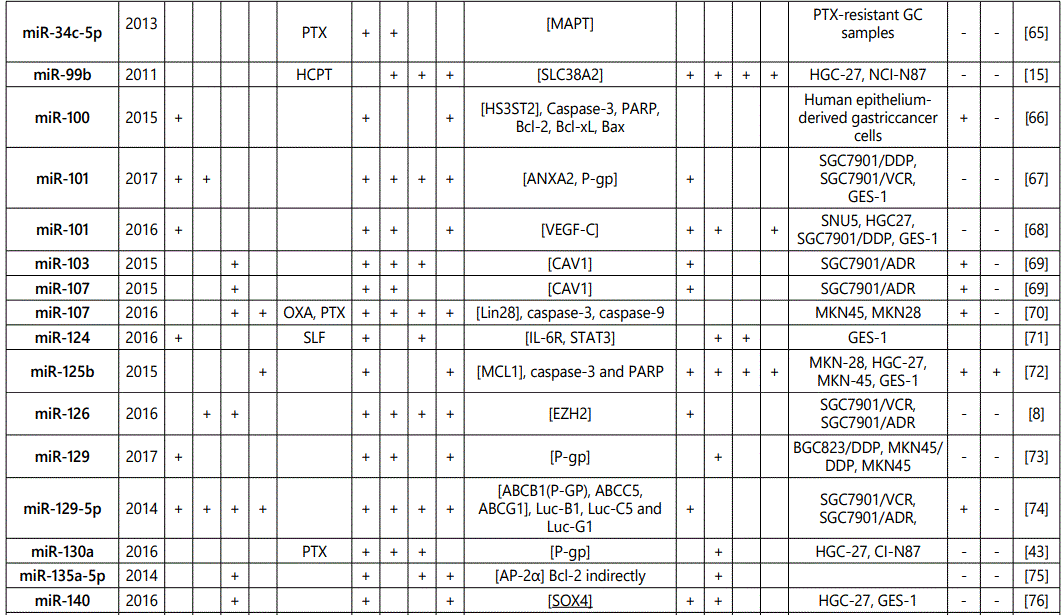
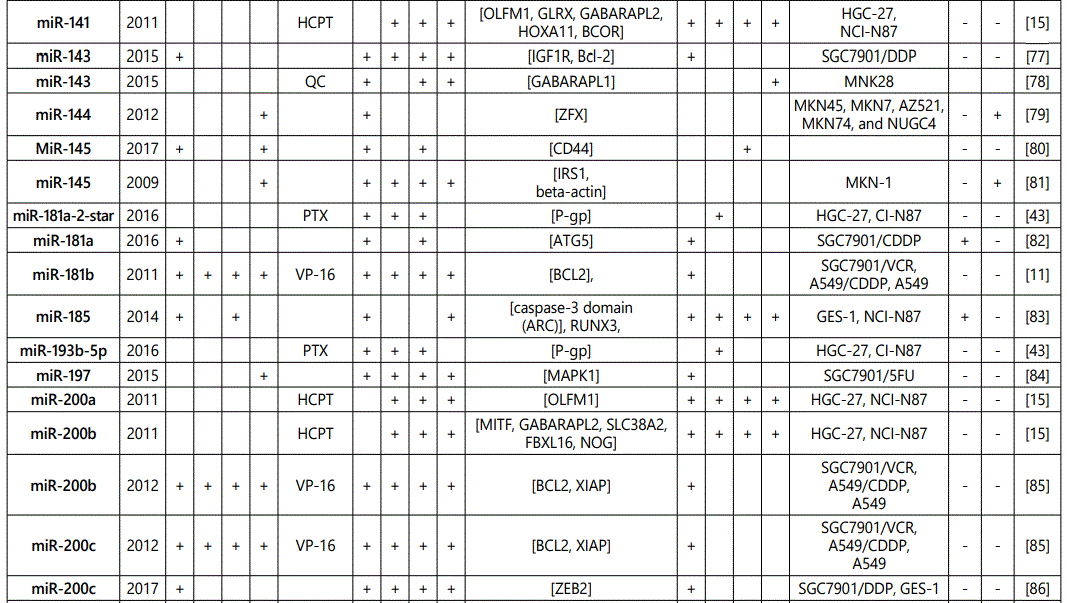
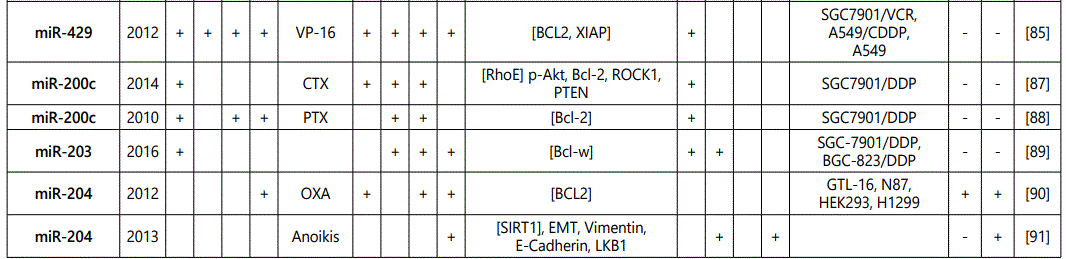
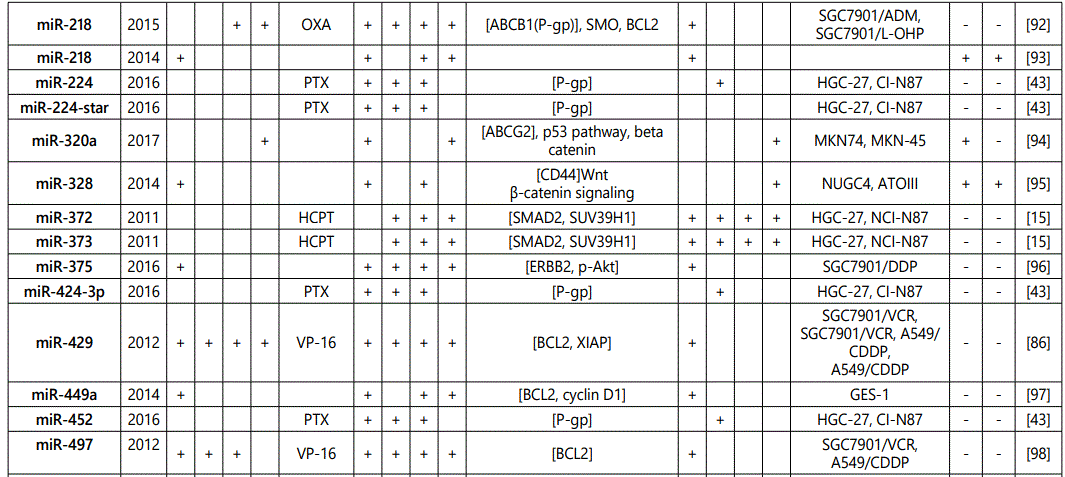
1Measurement methods for defining chemo-response (A-D): A: Growth rate/proliferation or colony formation; B: differential expression in resistant vs. parental GC cell lines; C: rate of growth inhibition by various drug concentration or IC50 detection; D: apoptosis rate (detailed information can be found in the methods section. *NOS: not otherwise specified. HCPT: Hydroxylcamptothecin; LPB: Lapatinib; QC: Quercetin; SLF: Sulforaphane; CTX: Cetuximab; DTX:Docetaxel; TSZ: trastuzumab; PTX: Paclitaxel; OXA: Oxaliplatin. Direct targets of the miRNAs are mentioned within [brackets]. Those out of brackets are other associated genes or pathways.
Direct targets of the mirnas are mentioned within [brackets]. Those out of brackets are other associated genes or pathways
miRNAs dysregulation modulates response to chemotherapy
Response to cisplatin: The list of miRNAs reportedly found dysregulated in resistant- vs. parental-GC cell lines can be found in tables 1 & 2. According to table 1, miR-223, miR-196, miR-92 and miR-181a represent the highest up-regulation in GC cell lines resistant to cisplatin vs. parental cell lines. The list of miRNAs whose up- or down-regulation had been associated, respectively, with significant decrease or increase in response to cisplatin therapy can be retrieved from table 3. Conversely, several other miRNAs have been shown if upregulated, they will decrease resistance to cisplatin therapy, and if down-regulated, on the other hand, they will show enhanced resistance (table 4). Figure 2 shows the trend by which cell viability changes in an environment of an increasing concentration of cisplatin, for GC cell lines transfected with either miRNA or antagomiR vs. control-vectors. Figure 3 shows differential IC50 concentrations detected from GC cell lines transfected with miRs/antagomiRs vs. controls. Studying IC50 concentrations gap, agomiR-26a had been found to induce the highest cisplatin sensitivity in GC cells, while on the other hand its antagomiR-(26a) modulates the highest resistance (figure 3.A.a, & 3.A.b, respectively). Moreover, analyzing differential apoptosis rates in GC cell lines transfected with miR/antagomiR vs. controls, the 4 miRNAs with the highest apoptosis induction rates due to cisplatin therapy were miR-200b, -497, -200c, and -429 respectively, all having the common target of proto-oncogene Bcl-2. Even antagomiR-135a-5p that regulates the 2nd highest resistance to cisplatin in GC cell lines does also conversely upregulate Bcl-2 (figure 4.A.a & table 4).
Response to adriamycin (doxorubicin): Only two miRNAs have been reported to be dysregulated in adriamycin-resistant GC cell lines, both of which conferring sensitivity to the drug (miR-126 & -101). Table 3 summarizes data of the studies listing miRNAs if overexpressed, they can increase resistance to adriamycin, and table 4 lists reports of miRNAs with exactly converse effects. Figure 2 plots trends by which cell viability changes for GC cell lines transfected with miRNA- (figure 2.B.a) or antagomiR- (figure 2.B.b) vs. control-vectors in the context of exposure to varying concentrations of adriamycin. Figure 3.B.a. lists the IC50 values for adriamycin in GC cell lines transfected with agomiRs vs. negative controls, and figure 3.B.b. shows the same data for antagomiR-transfected GC cells. A precise look at the latter two figures (3.B.a & b) shows that all the three miRNAs inducing the highest resistance to adriamycin in GC cell lines (-19a, -19b, -106a) have a common target of PTEN, while all the three antagomiRs (-200c, -218, -497) conferring highest sensitivity to doxorubicin target Bcl2. Figure 4.C shows the list of miRNAs that if transfected into a GC cell line can modulate adriamycin resistance. There is similarity between apoptosis and IC50 modulators of adriamycin. RUNX3 has been reported as a target for only two miRNAs in all the reviewed studies in this systematic review (-106a, 185), both modulating GC response to adriamycin by modulating, respectively, IC50 elevation (resistance) and enhanced apoptosis induction (sensitivity).
Response to 5-fluorouracil (5-FU): The list of miRNAs dysregulating the 5-FU resistant GC cell lines are summarized in tables 1 & 2. Tables 3 & 4 summarize chemoresponse to upor down-regulation of several miRNAs performed through transfection with agomiRs or antagomiRs, respectively. Figure 2.C. plots cell viability rates in GC cell lines transfected as abovementioned in the context of 5-FU therapy. The chemomodulation associates of 5-FU determined through differential response in IC50 levels or apoptosis rates are not very specific and is quite similar to that of adriamycin, suggesting existence of a cross-response for the two agents in GC cell lines (Figures 3.C & B and 4.C & D). A strange observation about 5-FU induced apoptosis by miRNA modulation was that, in one study [57], transfection by either agomiR- or antagomiR-30a, both induced resistance to apoptosis (figure 4.D.a & b).
Response to vincristine: Table 2 lists the miRNAs downregulated in the chemo-resistant GC cell lines, among which, those regulating resistance to vincristine therapy represent the lowest expression. miR-15b, miR-33a, miR-220, and miR-612 all represent less than 30 times expression in the vincristine-resistant GC cell lines versus the parental cells. As can be simply conceived from figure 3.D, no miRNA had been reported modulating increased IC50 doses for vincristine vs. controls in GC cells. Among miRNAs with the lowest IC50 modulation for agomiRs (miR-429 and -27b), both of them target Bcl-2. The highest vincristine IC50 were found for antagomiR-27b (targets CCNG1 & dysregulates p53) & -23b-3p (targets: ATG12 & HMGB2) transfected GC cell lines.
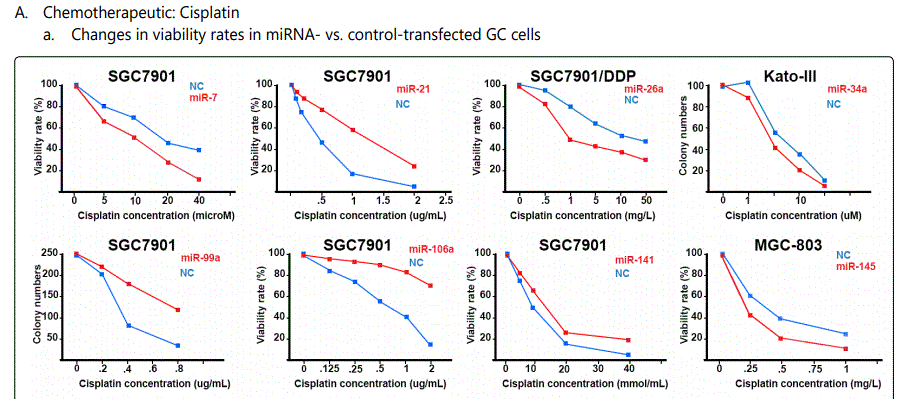
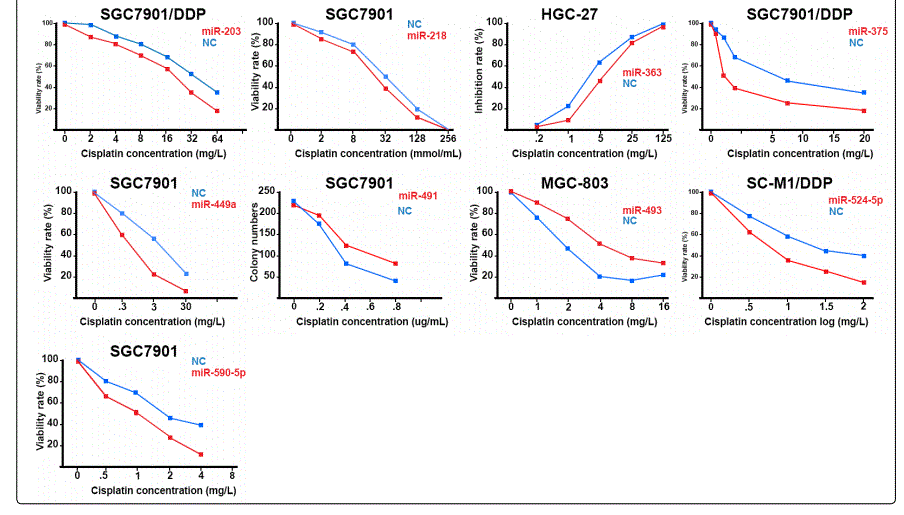
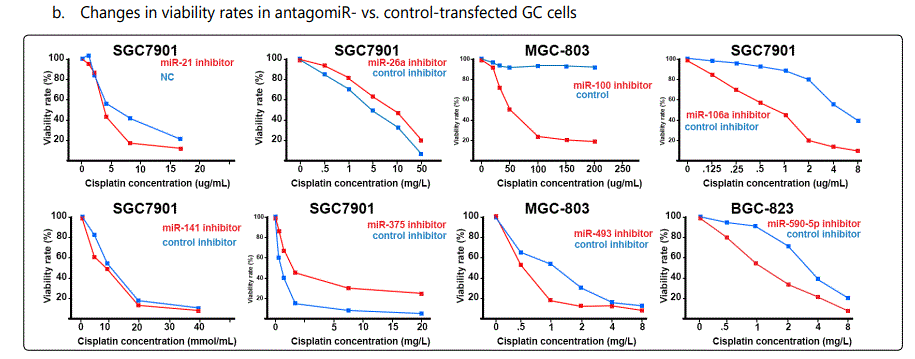
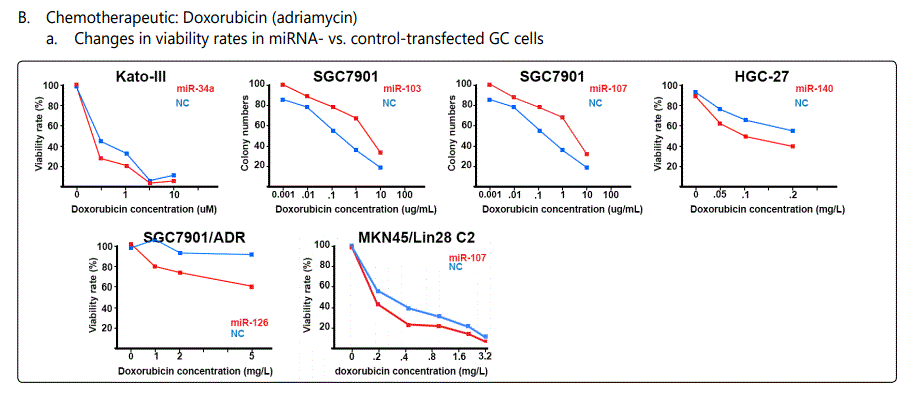
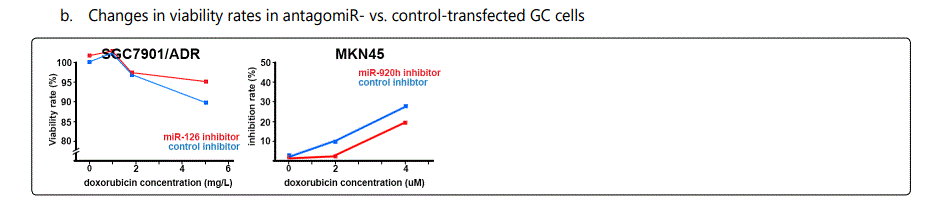
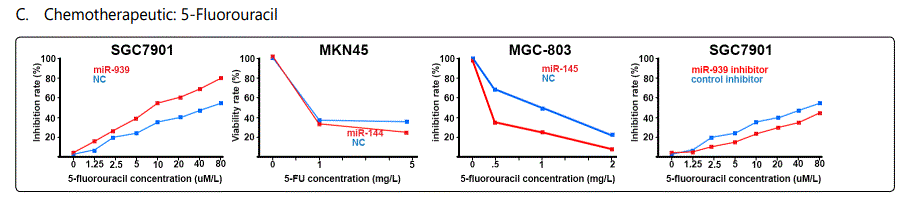
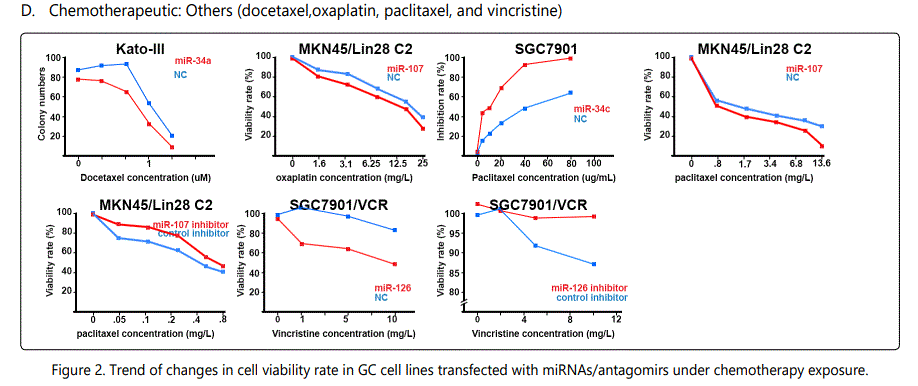
Foot note: Data used for plotting figure(s) 2 have been derived mostly from charts and figures presented at the studies, and so, the measurements are based on estimations of those charts; the SD part has also been excluded from the figures;
*Reports have been arranged in the decreasing order for chemo-resistance induction for agomiRs and chemo-sensitizing trend for antagomiRs; figures associated with non-significant difference have also been shown.
Controversial Reports
For some of the miRNAs, controversial reports existed in the reviewed studies, and while they were conferring chemoresistance or –sensitivity to the GC cell lines in one study, in another they were either representing no significant impact or an adverse to the former. Here is the full list of miRNAs for which different studies reported controversial effects:
miR-16: While in one study, miR-16 had been reported to be over-expressed in either SGC7901/DDP or BGC-823/DDP versus their parental cell lines [9], in four studies [12, 46-48] it had been associated with chemo-sensitivity inductions. Nonetheless, it is very important to notice that in none of the four studies mentioned, cisplatin resistance had been investigated, and cell lines resistant to Adriamycin, vincristine, and/or trastuzumab had been used. A simple explanation could be that miR-16 could confer chemoresistance to cisplatin but it offers chemo-sensitivity to the other chemotherapeutic agents, above mentioned.
miR-20a: miR-20a has been reportedly associated with chemoresistance in three studies [16-18] but it had been down-regulated in chemoresistant cell lines in one another [12]. Again, a more precise review of data in these studies reveals that in the latter study, SGC7901/VCRcell line had been used, while in two of the former ones, the cell lines used were resistant to cisplatin (SGC7901/DDP) and in the third one, sensitive cells had been exposed to cisplatin or docetaxel. Therefore, the simplest explanation for the observed controversy could be again a disparity in inducing either chemo-resistance or –sensitivity by the respected miRNA to different agents.
miR-21: Six studies [19-24] reported associations between miR-21 and chemoresistance to cisplatin, trastuzumab or paclitaxel while one another study [9] reported no differential expression of miR-21 in cisplatin-resistant cell lines versus parental cells.
miR-31: miR-31 was reported to be upregulated in SGC7901- 5FU cell line versus parental cells in a study by Wang et al. [6], suggesting a role as resistance inducing microRNA to 5-fluorouracil (and probably also CDDP, VCR, & ADR, because the cell line represented higher IC50 for all of the mentioned agents versus the parental SGC7901 line), while in a recent study, miR-31 had been reported as a chemo-sensitizing microRNA to 5-fluorouracil [58]. No explanation could be made based on the existing data, but future studies with more controlled approaches are recommended.
miR_106a: While three studies [7, 29, 30] reported miR-106a as a resistance conferring miRNA to multiple drugs (including its up-regulation in SGC79-1/VCR cell line), one another study [12] reported its downregulation in SGC7901/VCR cell line versus parental cells. Therefore, future studies are recommended.
miR-107: Two studies reported miR-107 as a positive multidrug modulator in GC cell lines to adriamycin, 5-fluorouracil, oxaliplatin and paclitaxel [69, 70], one study [9] reported its upregulation in either SGC7901/DDP or BGC-823/DDP versus their parental cells. The disparity in the chemotherapeutic agentsunder investigation suggests that miR-107 might have differential effects in chemoresponse to different agents.
miR-126: miR-126 had been reported as a modulator of chemo-resistance in one study [15] and chemo-sensitivity in another [8]. Again, there was a disparity in the chemo-agent under study, and miR-126 had been reportedly conferring chemoresistance to hydroxycamptothecin but chemosensitivity to VCR and ADR.
miR-141: miR-141 was reported as a modulator of chemoresistance to cisplatin [32] and chemosensitivity to hydroxycamptothecin [15].
miR-181a: Two studies reported miR-181a as a chemosensitivity predictor for paclitaxel [43] and cisplatin [82] therapy, while another study reported its upregulation in SCG7901/DDP and BGC-823/DDP. Further studies would be needed for elucidations.
miR-196a: One study reported miR-196a as a modulator of GC cell lines resistance to hydroxycamptothecin while another study reported its downregulation in SGC7901/VCR compared to the parental cells. Again, the disparity in the agent under investigation suggests a differential impact of miR-196a in different chemotherapeutic settings.
miR-224: One study [43] proposed miR-224 to confer chemosensitivity to paclitaxel in GC cell lines, while in another one [15] it has been associated with chemo-resistance induction to hydroxycamptothecin.
miR-452: There are two studies on the modulation effects of miR-452 in GC cell lines: One suggesting it as a resistance indicator to hydroxycamptothecin [15], while another one associated it to increase chemosensitivity to paclitaxel. Further studies in different aspects of the observed disparity, including on the metabolic pathways through which these two chemotherapeutic agents work, may to some degree explain the identifieddifference.
miR-let-7g: Two studies reporting controversial effects for miR-let-7g have been found in this systematic review. While in one study it has been associated with chemo-resistance to hydroxycamptothecin therapy [15], another one suggests it as a positive modulator of sensitivity to radiotherapy in GC cells lines [113].
Difference observed between subtypes of particular microRNAs: There are reports in which subtypes of some specific miRNAs conferring different effects on chemotherapy efficiency in GC cell lines. Here is a list of them: miR-196 vs. miR-196a; miR-181a vs. miR-181c\d; miR-106a vs. miR-106b; miR-99a vs. miR-99b; miR-17 vs. miR-17-5p; miR-23a vs. miR-23b; miR-190 vs. miR-190b; miR-193-5p vs. 193-3p; miR-424 vs. 424-3p; miR-let7g vs. miR-let-7a& miR-let-7b.
Discussion
Gastric cancer is an insidious malignancy that develops and progresses very silent, and in most cases it gets diagnosed in just very late stages, and despite all the preventive and screening strategies developed, it is still associated with very poor prognosis in reports from all over the world [114]. So, development of individualized treatment strategies is extremely relevant, and to attend this obligation, there have been substantial efforts with a large number of publications in this regard in recent years [59]. This systematic review, however, aims to attend the issue in the most comprehensible method, in order to make a well landscape of the existing knowledge on the issue, and provide a broad prospect for future research directed at individualization of therapy in GC.
Although a very large list of miRNAs has been shown to significantly dysregulate response to chemotherapy, it seems more logical to find and concentrate on those miRNAs that repetitively have been associated with the highest impact on response to chemotherapy, by different studies.
miRNAs most differentially expressed in chemo-resistant vs. parental cell lines
Detailed data of miRNAs highly dysregulate in the chemoresistant GC cell lines can be found in tables 1 & 2, or the Results section. The miRNA with the single highest gap of expression level was found to be miR-612 with more than 43 times down-regulation of the miRNA in vincristine-resistant SGC7901 cells vs. the parental cells, followed by miR-33a (≈36 times) and miR-15b (≈34 times). In fact, such large gaps of miRNA expression rates were not found for other drugs than vincristine. For cisplatin the largest gap was about 6.5 fold expression of miRNA-1301 modulating DDP-resistance. On the other hand, for doxorubicin, miR-126 represented 5 times less expression in ADR-resistant cells. Finally, for 5-FU, miR-22 represented the single highest magnitude with about 4 times less expression rate in 5-FU resistant cells than the parental cells.
miRNAs most considerably affecting IC50
Among miRNAs affecting cisplatin IC50, miRs-20a, -106a, -363, & -19a were associated with the highest differential IC50 indicative of resistance to cisplatin, while miRs-26a, -181a, -181b, and -23b-3p conferring lowest IC50 gap, and conferring cisplatin sensitivity (figure 3.A). For adriamycin, miRNAs -19a, -19b, -106a, and, -34a represented highest IC50, and -429, -218, -16-1, and -181b showed the lowest IC50 (figure 3.B). 5-fluorouracil IC50 most heavily raised due to overexpression of miRNAs -19a, -106a, 363, and -19b, and declined most profoundly by miR-218, -let-7b, -30a, and -27b (figure 3.C). Vincristine IC50 was raised by overexpression of miR-429, -27b, and -200c, and down-expression of miR-27b, 23b-3p and let-7b (figure 3.D).
miRNAs affecting apoptosis rates at the highest magnitude
Figure 4 plots differential apoptosis rates induced by 4 main GC chemotherapeutic drugs in the presence of over- or –under-expression of miRNAs. miRNAs most significantly enhance cisplatin-induced apoptosis include miR-200b, -497, -200c, -429, and -185, and those decrease apoptosis include miR-99a, -491 (detected by antagomiR study) and -362 (agomiR transfection). Vincristine induced apoptosis has been most significantly enhanced by upregulation of miR-15b and -16. Doxorubicin induced highest rate of apoptosis after overexpression of miR-218 and -27b, and lowest by miR-645. Finally, miR-15b overexpression was associated with the highest rate of 5-FU induced apoptosis rate in GC cell lines.
Common targets for miRNAs modulating resistance to chemotherapy
miRNAs induce their effects through down-regulation of their target mRNAs by attaching their 3' UTR. Nonetheless, this direct effect in most of the time activates networks of biological pathways that results in either up- or downregulation of several important factors playing significant roles in the biological effects induced by overexpression of an miRNA. In this section, the important genes and pathways that have been reported to be dysregulated by the expression of the miRNAs associated with response to chemotherapy in GC, have been reviewed. Most of the reviewed genes or pathways in the current systematic review have been reportedly as a target to both types of miRNAs that either induce chemo-resistance or those conferring chemosensitivity. The reason behind such a controversy lies in the disparity of different miRNAs effect in inducing either down-or up-regulation of a particular mRNA or biological pathway. Below some of the most important target mRNAs, genes or biological pathways that are directly or indirectly dysregulated by the reviewed miRNAs have been discussed. The miRNAs which are within [brackets] have been reported to directly target the 3' UTR of the mentioned gene or pathway, and inevitably a down-regulator of the mRNA, while those without brackets are due to reports of dysregulation of the target genes by the mentioned miRNAs (whose effect could be either down- or up-regulation). Regulation of the target genes by the mentioned miRNAs (whose effect could be either down- or up-regulation).
FBXW7 (F-box and WD repeat domain-containing 7) isthe substrate recognition component of an evolutionarily conservedSCF (complex of SKP1, CUL1 and F-box protein)- typeubiquitin ligase complex, which has been shown to have important roles in regulating multiple oncoprotein substrates, including cyclin E, c-Myc,Notch, c-Jun, mTOR and MCL1 [115]. Table 1 shows that in cisplatin-resistant cell lines, the miRNA (-223) up-regulated at the highest rate targets FBXW7. Similarly, miR-363 which represents the 3rd highest IC5O for cisplatin also targets FBXW7. It proposes FBXW7 as a common target for miRNAs that regulate resistance to cisplatin. Although there would be need for future studies to confirm this finding, if this confirmation happens anyway, its importance would be very high. The least imaginable relevance of such findings is that, before starting chemotherapy in a GC patient, practitioners instead of screening a large number of miRNAs to decide which chemotherapy to use, they can simply use the target levels of the respective miRNAs to predict the resistance status of a particular GC tissue to an agent; and in the case of cisplatin, if analysis of a patient's tissue showed down-regulation of FBXW7, it can be highly regarded as a cisplatin-resistant GC, and initiation or switching to alternative chemotherapeutics will be recommended.
PTEN is a tumor suppressor gene that had been a direct target of some of the miRNAs that most powerfully modulate chemo-resistance. This systematic review found 8 miRNAs that specifically target PTEN in their pathways related to GC. They include [miR-19a], [miR-19b], [miR-21] (4 studies), [miR-106a], [miR-193-3p], miR-200c, [miR-221], and [miR-222]. Expression of the first 5 miRNAs have been associated with resistance to chemotherapy, while the last three were conferring resistance to radiotherapy (tables 1,2,3,5). PTEN is the target of miRNAs whose down-regulation in GC cell lines modulate resistance to cisplatin, adriamycin, 5-FU, paclitaxel, and trastuzumab. No data was found about any potential effect of PTEN dysregulation and response to vincristine or docetaxel.
Bcl-2 is an anti-apoptotic gene whose expression is dysregulated in several cancers. Five miRNAs with chemo-resistance effects reported to affect Bcl-2 levels include: miR-19a, miR-19b, miR-27a, miR-362(Bcl2L1), and miR-503 (table 3); while 21 miRNAs with chemo-sensitivity induction are: [miR-15b], [miR-16], [miR-27b], [miR-34a], miR-100, miR-135a-5p, [miR-143], , [miR-181b], [miR-190b], [miR-200a], [miR-200b], miR-200c, [miR-429], [miR-200c], [miR-204], miR-218, [miR-429], [miR-449a], [miR-497] (2 studies), [miR-503](2 studies), miR-508-5p, [miR-1271] (table 4); and [miR-190b] is reported to be involved in inducing radio-resistance (table 5); three miRNAs although directly target Bcl-2, they reportedly confer no effect on chemo-response include: [miR-181a] [miR-181c], [miR-181d] (table 6). Bcl-2 is the target for miRNAs inducing resistance to cisplatin, adriamycin, vincristine, gemcitabine, docetaxel, and etoposide, while mitomycin sensitivity of GC cell lines showed no significant dysregulation after transfection with miR-181s that their target is Bcl-2. Similar finding was detected for 5-FU, although in one another study [6], up-regulation of Bcl-2 had been associated with resistance to 5-FU, suggesting that in GC cell lines with Bcl-2 up-regulation, the best choice can be mitomycin.
MDR1 encodes P-glycoprotein (P-gp) which is an energy-dependent efflux pump of ATP-binding cassette (ABC) transporters that could export planar hydrophobic molecules including some chemotherapeutic drugs (such as cisplatin,doxorubicin, vincristine, vinblastine, paclitaxel, colchicine, actinomycin D and mitomycin C) and consequently decreasethe intracellular level of drug accumulation; and so act as a chemo-resistance gene. There were 11 different miRNAs that reportedly induce chemoresistance and also disregulate MDR1/P-gp pathway: miR-10b, [miR-21], miR-22, miR-27a (2 studies), miR-31, miR-133b, miR-190, miR-501, miR-501-5p, miR-615, miR-615-5p (table 3). On the other hand, 37 miRNAs also reported to target MDR1/P-gp pathway in order to induce chemo-sensitivity in GC cells: [miR-15b], [miR-16], [miR-23b], [miR-27b], miR-30a (2 studies), miR-32, [miR-101], [miR-129], [miR-129-5p], [miR-130a], [miR-181a-2- star], [miR-193b-5p], miR-197, miR-201, [miR-218], [miR-224], [miR-224-star], [miR-424-3p], [miR-452], miR-486-3p, [miR508-5p], [miR-582-5p], miR-532, miR-766, miR-877, miR-1224-3p, miR-1229, miR-1238, miR-1273d, miR-3131, miR-3149, miR-3162-3p, miR-4701-3p, miR-4728-3p, miR-4763-3p, and miR-5096 (table 4). As it can be well conferred from its name, GC cell lines with upregulated miRNAs targeting MDR1 show resistance to all of the studied chemotherapeutic drugs. So, if future studies can find some drug that their effect are not affected by MDR1 dysregulation may revolutionize the treatment strategy of GC.
MCL1 or myeloid cell leukemia 1 is a pro-survival member of the Bcl-2 family, whose expression is supposed to be associated with tumorigenesis, invasion and resistance to chemotherapy. In the current review research, a total of 7 miRNAs associated with chemo-response, have been reported to dysregulate MCL1 expression. MiR-223 is the only miRNA in the chemoresistant group that had been reported as a downstream target of miRNA. [miR-17-5p], [miR-20a], [miR-106a], [miR-106b], [miR-125b], and [miR-320] have been reported as directly targeting MCL1 as their pathway in inducing chemo-sensitivity. The current systematic review showed GC cell lines miRNAs that dysregulate MCL1 can modulate resistance to trastuzumab and docetaxel. There is also indirect data (miR-106a effects reported by other studies) that says it can also compromise response to cisplatin, adriamycin and 5-FU, as well. Nonetheless, no association with response to vincristine and MCL1 dysregulation was found.
Caspase-3 is an important effector in pro-apoptotic pathways, and so is usually considered as a modulator of chemo-sensitivity in cancers. In the current systematic review, five miRNAs that have been reported to enhance chemo-resistance in GC cells dysregulate caspase-3 pathways: miR-19a, miR-19b, miR-421, miR-645, and miR- BART20-5p. 10 more miRNAs among the chemo-sensitive modulators have also been reported to significantly dis-regulate caspase-3 expression: miR-15b, miR-16, miR-27b, miR-34a, miR-100, miR-107, miR-125a, [miR-185], miR-508-5p, and miR-939. Cisplatin resistance has been reported by caspase-3 inactivation through several miRNAs. miRNAs activating caspase-3, on the other hand, enhanced sensitivity to cisplatin, 5-FU, adriamycin, vincristine, and etoposide. Caspase-3 dysregulation seems not to affect response to mitomycin C in GC cell lines [11].
p53 is an important denominator of the G1-S cell cycle checkpoint and a strong pro-apoptotic modulator that plays pivotal roles in preventing neoplastic transition in several cancers including gastric carcinoma. The current study found 8 miRNA whose expression levels alter p53 regulation in GC cell lines, 6 of which in the chemo-sensitizing miRNA group: miR-23b, miR-27b, miR-34a, miR-320a, miR-4496, and miR-508-5p (table 4). The remaining 2 miRNAs are involved in radio-sensitivity modulation: miR-195 and miR-503 (table 5). No resistance through down-regulation of p53 by any miRNA had been found, but response to cisplatin, vincristine, 5-FU, adriamycin, and even radiotherapy was found to be enhanced in GC cell lines in which p53 is up-regulated.
Akt pathwaysare well known cell survival pathways and its activation enhances resistance to apoptosis, and also is a reported indicator of resistance to chemotherapy in cancer cells. Overall 13 miRNAs reviewed in this study have been reported to target Akt pathway in some way, from which 9 miRNAs were modulators of chemo-resistance: miR-19a, miR-19b, miR-21, miR-106a, miR-590-5p, miR-942, miR-1287, miR-3127-5p, and miR-4713-5p; and 4 of miRNAs conferring chemo-sensitivity: [miR-29b], miR-34a, miR-200c, and miR-375. Overexpression of Akt in the GC cell lines, regulated by several miRNAs, had been reported to enhance resistance to cisplatin, adriamycin, 5-FU, trastuzumab, paclitaxel, and TRAIL treatment [6, 23, 29, 42, 43]. No data on its potential effects on vincristine or docetaxel were found; future research on this is therefore recommended.
STAT3 is constitutively an effectors of cell survival in association with surviving expression in cancer cells.Six miRNAs have been found associated with STAT3 dysregulation from which one (miR-590-5p) was among chemo-resistance promoters and the remaining 5 were modulators of chemo-sensitivity: [miR-17-5p], [miR-20a], [miR-106a], [miR-106b], and [miR-124]. Paclitaxel resistance has been associated with STAT3 down-regulation by miR-590-5p overexpression [40].
DNA repair pathways are one of the most important pathways that affect cancer result in GC. Among all the molecules and genes in this pathway, PARP1 was the only DNA repair pathway associated with chemo response alteration by miRNA expression in GC cell lines. The miRNAs acting through alterations of PARP1 pathway in chemo response include miR -21, miR-99a, miR-223, miR-421, miR-491, miR-BART20-5p, miR-100, miR-125b, and miR-939.
Limitations
Findings of the current systematic review is based on measurements reported by different authors from different studies, and despite the overall similar approach they followed, existence of some disparities in different studies is not dismissible. So, in order to have a more accurate evidence to make a decision-making algorithm in the treatment of GC, there would be need for prospective studies that mention all the data retrieved from this systematic review in a single well-designed comprehensive study. Also, even when studied by a unique study, there could be large gap in the observations of one manipulation in different cell lines. For example, miR-34a transfection into Kato III GC cell line induced 2.4 times larger IC50 vs. controls, compared to only about 1.2 times when the same transfection performed in MKN-45 line in one study [61]. So, there are a large number of factors, other than miRNAs, significantly interfering in the development of resistance or sensitivity to chemotherapy in GC, which needs to be well controlled, before making any final judgment. There was no such a possibility in the current study, but its findings can serve as a commencement point for those who want to further investigate in this field in order to confirm or revise the findings of the current study.
Conclusion: Approach to a decision-making algorithm for GC therapy
There had been efforts in the literature to make a signature of miRNAs or other genetic and epigenetic factors, to provide a targeted approach to therapy in GC patients. Nonetheless, most of these efforts were based on merely surmises, and there were no indicative data before to guide conducting these studies. This study, however, provides invaluable data about some potential markers that can be used in this regard. So the question is, can we construct a reliable targeted therapeutic approach based on the findings of the current systematic review?
In order to effectively improve the effects of chemotherapy in inhibiting GC progression and inducing apoptosis to GC cells, there is a need to develop an algorithm that uses the most significant correlates of response to chemo-drugs in GC. Nonetheless, the existing data is highly limited and future directed researches are needed to explore the escape doors in tough situations, when no therapy seems to be effective. For example, in the current systematic review, it has been demonstrated that in the context of an elevated Bcl-2 in GC cell lines, cisplatin should be avoided because all the miRNAs with the highest rate of cisplatin-induced apoptosis have the common target of Bcl-2; and the only choice of treatment could probably be mitomycin in this case. On the other hand, if Bcl-2 is detected to be low I the tissue, the best treatment is adriamycin (or vincristine), because the three miRNAs modulating adriamycin sensitivity all have the common target of Bcl-2. Nonetheless, if PTEN is down-regulated, adriamycin is dismissible, because all the 3 miRNAs most heavily regulating adriamycin sensitivity commonly target PTEN. And if the feasibility of such algorithms is confirmed by prospective researches, or after making revisions anyway, then commercial kits can be developed which could effectively subside the prices and make it an acceptable standard approach to GC therapy in clinical setting, worldwide.
Acknowledgement
Author would like to acknowledge Mr. Morteza, Mr. Rahmat and Ms. Negar Taheri Amlashi, with special thanks for their kind help to the Encyclopedia Amlashica.
Conflicts of Interest
The author has no conflicts of interest to declare.
References