Research Article
Quantification of Rabies Virus by Real Time PCR in comparison with Mouse Inoculation Test (MIT) and Fluorescent Antibody Test (FAT)
Pasteur Institute of India, Coonoor, The Nilgiris, Tamilnadu, India
*Corresponding author: Thangaraj Sekar, Assistant Research Officer, Pasteur Institute of India, Research and Development, Coonoor, The Nilgiris, Tamilnadu, India, Tel: +9487365795, E-mail: sekar_102000@yahoo.com
Received: March 11, 2019 Accepted: March 19, 2019 Published: March 25, 2019
Citation: Sekar T, Premkumar AA, Mohan GC, Sekar B, Sundaran B, Sivakumar S. Quantification of Rabies Virus by Real Time PCR in comparison with Mouse Inoculation Test (MIT) and Fluorescent Antibody Test (FAT). Madridge J Vaccines. 2019; 3(1): 80-85. doi: 10.18689/mjv-1000118
Copyright: © 2019 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Real time PCR is the modern molecular techniques used for detection and quantification of viral particle in the specimens. In this study Real Time PCR (RT-PCR) based method was developed and used to quantify the rabies virus PV-11 strains in the different viral harvests of tissue culture fluid in comparison with the Mouse Inoculation Test (MIT) and Fluorescent Antibody Test (FAT). The Eagleʼs MEM tissue culture medium with 5% fetal calf serum, 1% neomycin and 0.5% Amphotericin-B were used for the propagation of Vero cells in 150 cm2 tissue culture flasks. The confluent monolayers of the Vero cells (90%) were infected with 0.2 MOI of PV-11 strain of rabies virus. Five individual viral harvests were collected from the infected vero cells at every 3 days interval and subjected for MIT, FAT to quantify the viral titre. The MIT titrated sample was used for the preparation of five point internal reference standard for RT-PCR technique to quantify the number of viral particles in the harvested fluid. The primers targeting 98 base pairs from 446 to 543 in the cDNA of the rabies virus nucleoprotein gene (N gene) were designed and synthesized for Real time PCR assay. The in-house RT-PCR assay was developed with SYBR green master mix. Viral particle in each harvests were quantified in comparison with internal reference standard included in the assay. The results of MIT, FAT and Real time PCR methods were compared with each other. The standardized Real time PCR method was applied for the quantification of viral titre in the different viral harvests cultured using Roller bottle and Bioreactor culturing methods using two different medium. Maximum log of 7.216 viruses per 1 ml of bioreactor viral harvest was quantified by using the real time PCR method. Dilution of the samples are required if the sample having higher viral titre which is not covered within the linear standard range of log 4.50 to log 6.50. This real time PCR based quantification of rabies virus has the advantage of greater accuracy, rapid, economical when compared to the other tests and no animal models required.
Keywords: Rabies virus; Mouse Inoculation Test (MIT); Fluorescent Antibody Test (FAT); Vero cells.
Introduction
Rabies virus is a most important zoonotic pathogen causes an acute encephalomyelitis associated with a high mortality in human and animals in the different part of the world, particularly in Asia and Africa [1]. The rabies virus belongs to genus Lyssavirus of the Rhabdoviridae family consists of single standard non-segmented negative sense RNA approximately 12 kb in size [2,3].
The rabies is a bullet shape virus with the length of 180 nm and diameter of 75 nm in size. The rabies virus encodes five proteins namely Nucleoprotein (N), Phosphoprotein (P), Matrix Protein (M), Glycoprotein (G) and Polymerase (L) [4] frequent gene is to develop assays to diagnosis.
Rabies can affect both domestic and wild animals. It spread to people through bites or scratches, usually via saliva, over 95% of human deaths occurring in Asia and Africa regions are due to animal bites. Every year, more than 15 million people worldwide receive post-bite vaccination against rabies [5,6].
Globally canine rabies causes approximately 59,000 human deaths over 3.7 million disability-adjusted life years (DALYs) [7]. The N gene is the most conserved region among the RABV genome which is the reason for this frequent use as targets for rabies virus diagnostic assays [1,8].
The currently used Gold standard methods used for rabies diagnosis, the Fluorescence Antibody Test (FAT) and the Mouse Inoculation Test (MIT) are time consuming and are highly variable in specificity and sensitivity in the quantification assays. In this connection, the use of the RT-PCR for rabies virus detection was proposed [9] which proved to be a suitable technique for rabies diagnosis by various researches [10-12]. RT-PCR assays are used for the detection of Lyssavirus RNA from the samples like saliva, nuchal skin biopsy, eye washes etc with low viral load [13].
Over the decades many studies have been published assessing Real-time RT-PCR assays for the detection of RABV and other related lyssa viruses via SYBR green and TaqMan probes [8,14,15] methods. The real time PCR is most accurate, reliable and sensitive method for the quantification of specific target. The real time PCR allow rapid one step quantitative detection, the detection limit of PCR assay was 10 RNA copies per reaction with inter and intra assay variation below 4% [15].
Materials and Methods
Cell substrate and virus strain
A cryo preserved vial of Vero cell substrate in the passage level 141 was revived and subcultured as per the standardized protocols followed in our laboratory. Five numbers of 150 cm2 tissue culture flasks with confluent monolayer of vero cells were propagated and used for rabies virus PV-11 strain propagation and used for the real time PCR standardization purpose.
Viral inoculation and harvesting of viral fluid
0.2 MOI of PV-11 virus was diluted in 10 ml of PBS and infected to five numbers of 150 cm2 tissue culture flasks having about 90% of the confluent monolayer of the vero cells. The infected tissue culture flasks were incubated at 37°C for 90 minutes to facilitate the virus adsorption. During the adsorption period the viral fluid in the tissue culture flasks were shaken at every 20 minutes intervals. The fluid was discarded from the flasks and the pre-warmed growth medium (37°C) was replenished. The virus infected tissue culture flasks were incubated at 37°C incubator until the completion of final harvest. The viral harvests were collected and pooled in every three day intervals under aseptic condition. The flasks were replenished with fresh medium after every harvest which was supplemented with 2% of fetal bovine serum, 1% Neomycin and 0.5% Amphotericin-B (Maintenance Medium). At the end of every viral harvests the pooled viral fluid were centrifuged for 10 minutes at 2,500 g for the removal of cellular debris and stored at -80°C for further viral titration work. These five single viral harvests were subjected for rabies virus quantification using MIT, FAT and Real time PCR.
Methods for viral titration
Mouse inoculation test (MIT): 150 Nos. of Swiss albino mice (Male and Female equally distributed) in the weight range of 11-14 grams obtained from Pasteur Institute of India, Coonoor breading facility after proper approval of Institutional Animal Ethical Committee (IAEC) and maintained as per the CPCSEA guidelines. All the five single harvests were subjected for MIT test to find out the rabies viral titration units. As per the WHO procedures [16], 0.2 ml of each viral harvest were serially diluted with 1.8 ml of PBS containing 2% FBS (10 fold dilution) up to 10-6 dilution in the identical sterile set of tubes. 30 µl of serially diluted samples from 10-2 to 10-6 dilutions of five single viral harvests were injected intracerebrally to each respective group consist of 6 mice. The observation were recorded on daily basis from 0th to 14th day for rabies specific symptoms, mortality and the viral titre calculated using Spearman Karber method.
Direct Fluorescent Antibody Test (FAT)
Neuroblastoma-2A cells revival and subculturing for FAT: The cryo preserved vial containing 4.5 million Neuroblastoma 2A cells was revived in one 25 cm2 tissue culture flask in Eagleʼs MEM containing 20% FBS, 10% TPB (tryptose phosphate broth) with 1% neomycin 0.5% Amphoericin - B antibiotic (Revival medium) as same as vero cell revival procedure without the application of Trypsin EDTA. The Neuroblastoma 2A cells were further subcultured using MEM medium containing 10% of FBS, 10% TPB (tryptose phosphate broth) with 1% neomycin, 0.5% Amphoericin-B antibiotic (growth medium) and used in Fluorescent Antibody Test.
Fluorescent antibody test procedure: The five single harvests were subjected for fluorescent antibody test to estimate the viral titre. 0.2 ml of each viral harvest were serially diluted with 1.8 ml of growth medium (10 fold dilution) up to 10-6. In a 96 well microtiter plate 100 µl of growth medium was added in all the wells as per the test layout and 100 µl serially diluted samples were added (8 replicate) in the separate row for each dilution from 10-2 to 10-6. Approximately 25,000 Neuroblastoma-2A cells were added under aseptic condition to each well and the plates were incubated at 5% CO2 incubator for 48 hours. After the incubation period, the plate was observed for the growth of Neuroblastoma 2A cells under the cell imager. The contents in the 96 well microtiter plates were discarded aseptically into disinfectant solution. The cells were fixed by the addition of 100 µl acetone buffer with PBS at a ratio of 7:3 (pre-cooled prior to addition) to each well followed by incubation at -20°C for 30 minutes. The acetone buffer was discarded, 100 µl of anti-rabies antibody FITC (Fluorescein isothiocyanate) conjugate in PBS (1:20) was added to each well and the plate was incubated at 37°C for 30 minutes. The plate gently washed with sterile distilled water and drop of mounting buffer was added to each well before observing under the fluorescent microscope for the positive and negative fluorescence [16]. The results were calculated by Spearman Karber method after ensuring the positive and negative controls passes its validation criteria.
Real time-PCR
RNA extraction and cDNA synthesis: The viral RNA from five single viral harvests were extracted using Nucleospin viral RNA II extraction kit according to the manufacturerʼs instruction (Macherey-Nagel NucleoSpin viral RNA isolation kit). The RNA was eluted using 50µl Nuclease free water. 1 µl of RNA from each samples were subjected to cDNA synthesis by using iSCRIPT cDNA synthesis Kit (BIO-RAD). 20 µl cDNA synthesizing reaction mixture contains 4 µl of 5x iScript reaction mix, 1 µl of iScript Reverse Transcriptase, 14 µl of Nuclease free water, and 1 µl of template RNA. The following conditions were maintained for cDNA conversion: priming at 25°C for 5 minutes, reverse transcription at 42°C for 30 minutes, RT inactivation at 85°C for 5 minutes, optimal setup hold at 7°C in applied bio system veriti 96 thermal cycler and stored at -80°C for future use.
Standards for RT-PCR: The sample containing the viral titre log 5.520 in 1 ml (Estimated by MIT) was used for the preparation of five point linear standards. The RNA isolated from this sample was eluted using molecular grade Nuclease free water and diluted to log 5.50. The internal reference standards were prepared using fivefold dilution representing the viral copy number. 10 µl, 5 µl of RNA was added (volume of Nuclease free water reduced accordingly) in the cDNA synthesis reaction mix to represent the log 6.50 and log 6.00 respectively. The cDNA was subjected for fivefold serial dilution (4:1) to get the subsequent reference standards up to log 4.50. The internal standards ranging from log 4.50 to log 6.50(5 point calibration) were included in theassay to quantify the rabies viral copy number present in each viral harvests.
Real time PCR assay: Real Time PCR assay was carried out using the Roche light cycler 96 instrument. 20 µl reaction volume which comprises 10 µl of 2X SYBR green real time master mix, 7 µl of nuclease free water, 1 µl of forward primer, 1 µl reverse primer and 1 µl of template for each reaction used for real time PCR assay.
Amplification was targeted to the nucleoprotein gene which is about 98 base pairs. The genes were amplified for 35 cycles with the denaturation: 94°C for 20 sec, Annealing: 54°C for 25 sec and Extension: 72°C for 30 sec. The amplification of the each standards, controls and samples were monitored in each cycle and the negative control base line value maintained lower than 0.02 until the end of 35th cycle. The R value, efficiency were calculated by the light cycler 96 software with five point internal reference standard amplification using the primer pairs designed for N gene of rabies virus. The CT value of the internal reference standards and controls to be monitored during each cycle of amplification from our subsequent assays is to verify the repeatability. The rabies viral cDNA copy number of each sample were calculated using RT-PCR light cycler 96 software in comparison with the CT value of the five point internal reference standard included in the assay procedure.
Application Real time PCR for rabies virus quantification
The study was conducted with the objective of finding the difference in viral titre in the viral harvests collected from Roller bottle and Bioreactor culturing methods with two different medium namely MEM Eagleʼs (Himedia Cat # AT047) and RPMI 1640 (Himedia Cat # AT028). The viral titration in the 4 viral harvests of all eight batches (two different culturing methods with two medium) were quantified by real time PCR for the comparative analysis of viral titre in response to the culturing method and medium used in this study.
Results and Discussion
This study was mainly focused on the quantification of rabies virus propagated in vero cell line. Propagation of rabies virus in the vero cells for the manufacturing of tissue culture anti-rabies vaccine is the crucial stage and itʼs directly influence the vaccine production capacity.
The viral titrations in five individual viral harvests were estimated using conventional methods like Fluorescent Antibody Test (in-vitro) and Mouse Inoculation Test (in-vivo). Any quantitative measurement requires standards to quantify the targets in the unknown specimen. In this study the viral titre arrived at MIT was used for the formation of internal reference standards for RT-PCR to quantify the rabies virus.
Viral infection & harvest
0.2 MOI of viral titre was used for the infection of vero cells and 5 individual viral harvests were collected at 3 days intervals. Moreover suspended cellular debrisʼs were observed on 5th viral harvest. The 5th viral harvest was collected only from the viral titre standardization batch from 150 cm2 flasks.
Viral titration by in-vivo mouse inoculation test
The titre of the rabies virus PV-11 strain (used for the production of rabies vaccine) in the cell culture fluid was estimated using conventional mouse inoculation test. There were five group (10-2 to 10-6:5 dilutions) of mice consist of 6 mice in each group inoculated intracerebrally. The mice were observed on daily basis until 14th day for the development of rabies specific symptoms, mortality and recorded the results. The mortality ratio of the mice is indirectly proportional to the dilution of sample (lower dilution shows higher mortality). In subsequent dilutions decreasing pattern of mortality in the respective group of mice were observed based on the concentration of virus in each dilution. The rabies viral titres (lethal dose 50/LD50) of five single viral harvests were calculated as per Spearman Karber method and the results were tabulated in table 1. Among the five single viral harvests the second viral harvest shows the higher viral titre log of 5.85 and the fifth viral harvest shows the lowest viral titre log of 4.68 per ml.
Direct fluorescent antibody test
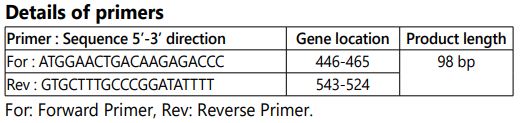
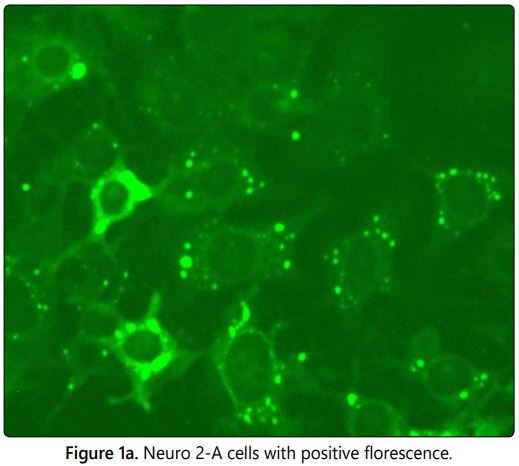
The fluorescent antibody technique was developed by Larghi et al in 1975 as an alternative method for the mouse inoculation test (MIT) since MIT has some disadvantages like time consuming procedure, use of live animal models and its requires ethical clearance. The Fluorescein isothiocyanate (FITC) labelled conjugated with rabies antibodies used to stain the PV-11 strain of rabies virus which were propagated in Neuro 2A cells at 96 well microtiter plate in the dilution ranging from 10-2 to 10-6. On observation under the fluorescent microscope the positive apple green colour fluorescence (Figure 1a) indicates the amplification of virus in the particular well and the absence of specific fluorescence (Figure 1b) in subsequent dilution indicates no viral particle in the sample (Negative/Endpoint). The positivity in the higher dilution is directly proportional to the concentration of virus in the sample. The normal cell control (negative control) was included in each test to rule out the cross contamination and to differentiate the non-specific florescence if any. The lower dilution shows the higher infectivity ratio by fluorescent foci and decrease in infectivity ratio (rabies specific florescence) in subsequent dilutions were observed in the Neuroblastoma 2A cells in correlation with the dilution of sample. The viral titres of the five single viral harvests were calculated (Infective dose 50/ID50) using Spearman Karber method and the results were tabulated in table 1. The results show that the second viral harvest has the highest viral titre of log 6.00 in 1 ml and the 5th viral harvest shows log of 5.125.
Rabies virus quantification by RT-PCR
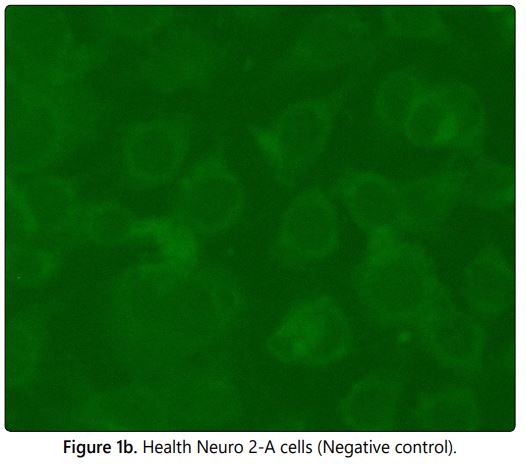
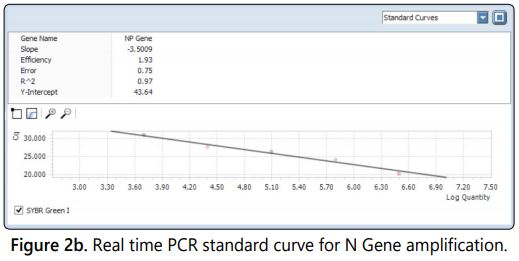
The real time PCR was standardized with the nucleoprotein (N) gene specific primer with the SYBR Green as a dye in the master mix. The gene sequence encodes for N gene is most conserved for rabies virus [17]. The internal reference standards ranging from log 4.5 to log 6.5 (5 standards) included in the test and the corresponding antilog value of the viral titre were calculated to viral copy number and used for the extrapolation of the viral copy number of unknown samples. The viral copy number and corresponding titre of all five viral harvests ranging from log 5.055 to log 5.961(Table 1) quantified by real time PCR absolute quantification using the standard curve. The fluorescence Curves of internal reference standards and samples were shown in figure 2a. A negative control was included in each test to ensure that there was no false positivity/viral titre raise due to cross contamination. The quantities of target gene in the samples were calculated from the difference of Ct (Cycle threshold) value between the sample and standards included in the assay [18]. The Ct values are inversely proportional to the copy number of the target gene in the reference standards/samples. The R value of the assay was 0.97 and the efficiency of the run was 1.93 calculated by the light cycler 96 software with five point internal reference standard included in the assay (Figure 2b).
Comparative analysis of viral titration in MIT, FAT & Reverse transcriptase RT-PCR
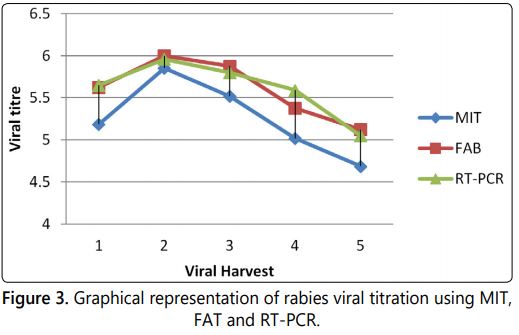
The results of MIT, FAT and reverse transcriptase RT-PCR for the five different harvests were tabulated below (Table 1) and the graphical representations of the same were shown in figure 3.
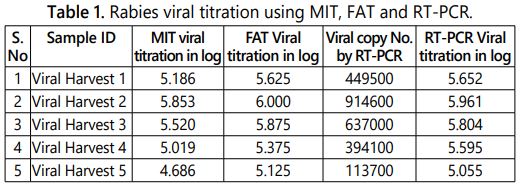
The RT-PCR and FAT based rabies virus quantification always shows the higher titre value than the MIT in each specific samples. The detection and quantification principle involved in the real time PCR is based on the fluorescence intensity produced in correlation with the viral RNA copy number in the sample. FAT detect the live amplified virus in the Neuroblastoma 2A host system by anti-rabies antibody tagged with FITC dye and the viral titre is calculated based on the ID50 value. The MIT viral titration based on the LD50 value of the sample observed in the injected animal model. The MIT and FAT only detects the live virus particles present in the specimen [19]. The inter assay variation between MIT to FAT was observed a maximum of 0.439 log and between MIT to RT-PCR was observed as maximum of 0.576 log.
Comparison of viral titration among roller bottle and bioreactor samples
The higher titre of the virus multiplication in the viral culturing method is directly proportional to the yield of vaccine in the vaccine manufacturing process. The factors like culturing methods (Roller culture or Bioreactor culture) and medium has the influence on viral titre during the large scale manufacturing of vaccine. 32 samples were analyzed by in-house standardized real time PCR viral quantification technique and the results were tabulated below (Tables 2a and 2b). It was observed that among the eight different batches the Bioreactor culture with MEM Eagleʼs medium has the higher rabies viral titre of log 7.216 per ml. The comparative analysis results of roller bottle and bioreactor culturing methods with two different media batches were tabulated in tables 2a and 2b.
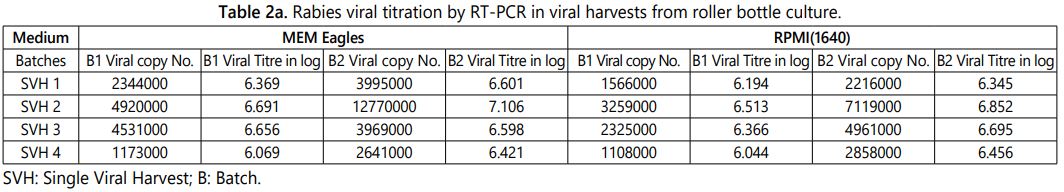
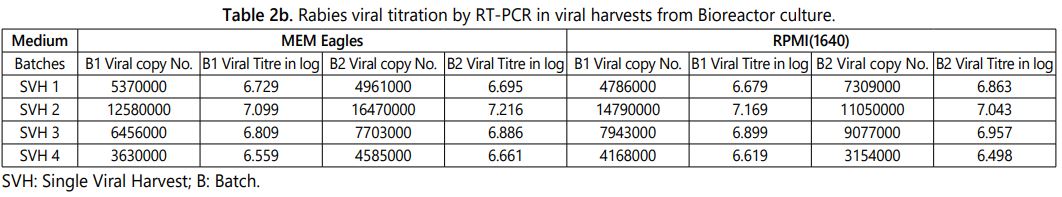
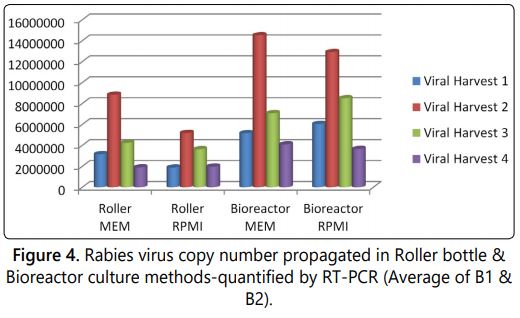
The above observation shows that the second viral harvest always has the higher viral titre than other three viral harvest and the same pattern were observed in the different batches irrespective of culturing method and medium (Tables 2a and 2b). The same pattern was observed in MIT, FAT and RT-PCR quantification methods while enumerating the viral titre among the five harvests propagated in 150 cm2 tissue culture flasks (Table 1). The roller bottle culture has the highest viral titre log 7.106 per ml (12770000 copy number) and the bioreactor culture has the viral titre log 7.216 per ml (16470000 copy number) of the MEM Eagleʼs medium viral harvest. The average of two batch results shows that the bioreactor culture has approximately 0.3 to 0.5 log higher viral copy number (titre) when compare to the roller bottle culture in the final harvested tissue culture fluid. Further very similar results were observed between MEM Eagles and RPMI 1640 tissue culture medium while culturing using same culturing method (Figure 4).
Conclusion
The Mouse inoculation test is the gold standard method conventionally used for the viral titration; it requires mice models and time consuming procedure. The Fluorescent Antibody Technique is a best alternative method for the quantification of the rabies virus and requires the suitable cell culture facility and trained laboratory technicians to handle the live rabies virus in the protocols. Now-a-days the molecular techniques such as real time reverse transcriptase PCR can be used for the rabies virus quantification in the viral harvest and the use of laboratory animal models can be minimized by replacing this with MIT. This preliminary attempt shows the encouraging results and it requires some more batch data to check the consistency of the results. Furthermore the RT-PCR can be used as alternative method i.e as an inprocess method for rabies virus quantification in the viral harvest fluids after fulfilling regulatory requirements.
Acknowledgement
The authors would like to thank the colleagues Mr. R Krishnamurthy, Mr. K. Ravichandran, Mr. M. G Subramani, Saravanamoorthy, Research and Development Laboratory and Mr. R.Gopalakrishnan, Mr. N.Chandran Quality Control Department of Pasteur Institute of India - Coonoor for their constant support and co-operation through the period of this project work.
References
- Fayea M, Dacheux L, Weidmann M, et al. Development and validation of sensitive real-time RT-PCR assay for broad detection of rabies virus. J Virol Methods. 2017; 243: 120-130. doi: 10.1016/j.jviromet.2016.12.019
- Bernardi F, Nadin-Davis SA, Wandeler AI, et al. Antigenic and genetic characterization of rabies viruses isolated from domestic and wild animals of Brazil identifies the hoary fox as a rabies reservoir. J Gen Virol. 2005; 86: 3153-3162. doi: 10.1099/vir.0.81223-0
- Deressa A, Pal M, Kobayashi Y, Kassa T. Molecular Epidemiology of Rabies Virus Isolates in Ethiopia. Int J Livest Res. 2015; 5(10): 21-33. doi: 10.5455/ijlr.20151027081633
- Dastkhosh M, Rahimi P, Haghighat S, et al. Cell culture extraction and purification of rabies virus nucleoprotein. Jundishapur J Microbiol. 2014; 7(9): e11734. doi: 10.5812/jjm.11734
- WHO Report. Rabies. World Health organization. 2018.
- Knobel DL, Cleaveland S, Coleman PG, et al. Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ. 2005; 83(5): 360-368.
- Hampson K, Coudeville L, Lembo T, et al. Estimating the Global Burden of Endemic Canine Rabies. PLoS Negl Trop Dis. 2015; 9(4): e0003709. doi: 10.1371/journal. pntd.0003709
- Hayman DT, Banyard AC, Wakeley PR, et al. A universal real time assay for the detection of Lyssaviruses. J Virol Methods. 2011; 177(1): 87-93. doi: 10.1016/j.jviromet.2011.07.002
- Bordignon J, Brasil-Dos-Anjos G, Bueno CR, et al. Detection and characterization of rabies virus in southern brazil by PCR amplification and sequencing of the nucleoprotein gene. Arch Virol. 2005; 150(4): 695-708. doi: 10.1007/s00705-004-0448-y
- David D, Yakobson B, Rotenberg D, Dveres N, Davidson I, Stram Y. Rabies virus detection by RT-PCR in decomposed naturally infected brain. Vet Microbiol. 2002; 87(2): 111-118.
- Dantas Junior JV, Kimura LMS, Ferreira MSR, et al. Reverse Transcriptionpolymerase chain reaction assay for rabies virus detection. Arq Bras Med Vet Zootec. 2004; 56(3): 398-400. doi: 10.1590/S0102-09352004000300017
- Gigante CM, Dettinger L, Powell JW, et al. Multi-site evaluation of the LN34 pan-lyssavirus real-time RT-PCR assay for post-mortem rabies diagnostics. PLoS One. 2018; 13(5): e0197074. doi: 10.1371/journal. pone.0197074
- Wadhwa A, Wilkins K, Gao J, et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay for the Detection of Highly Variable Rabies virus and Other Lyssaviruses. PLoS Negl Trop Dis. 2017; 11(1): e0005258. doi: 10.1371/journal.pntd.0005258
- Suin V, Nazé F, Francart A, et al. A Two-Step Lyssavirus Real-Time Polymerase Chain Reaction Using Degenerate Primers with Superior Sensitivity to the Fluorescent Antigen Test. Biomed Res Int. 2014; 2014: 256175. doi: 10.1155/2014/256175
- Coertse J, Weyer J, Nel LH, Markotter W. Improved PCR methods for detection of african rabies and rabies-related lyssa viruses. J Clin Microbiol. 2010; 48(11): 3949-3955. doi: 10.1128/JCM.01256-10
- Meslin FX, Kaplan MM, Koprowski H. Laboratory techniques in rabies. 4th edition. World Health organization, Geneva, (GN). 1996: 80-93.
- Jackson A, Jackson A, Wunner WH. Rabies. Academic Press: 2002; 322-334.
- Vaerman JL, Saussoy P, Ingargiola I. Evaluation of real time PCR data. J Biol Regul Homeost Agents. 2004; 18(2): 212-214.
- Franka R, Svrcek S, Madar M, et al. Quantification of the effectiveness of laboratory diagnostic of rabies using classical and molecular-genetic methods. Vet Med (Praha). 2004; 49(7): 259-267.


