Clinical Image Article
Immune Responses Induced by Candidate Optimized HIV DNA Vaccine in Phase I Clinical Trial
1State Research Institute of Highly Pure Biopreparations; The Biomedical Center; Peter the Great St. Petersburg Polytechnic University, Saint Petersburg, Russia
2Pavlov State Medical University, Saint Petersburg, Russia
*Corresponding author: Ekaterina Akulova, State Research Institute of Highly Pure Biopreparations, The Biomedical Center, Saint Petersburg, Russia, E-mail: ekaterina.akul@gmail.com
Received: May 21, 2017 Accepted: July 14, 2017 Published: July 20, 2017
Citation: Akulova E, Murashev B, Nazarenko O, et al. Optimized HIV DNA Vaccine in Phase I Clinical Trial. Madridge J Vaccines. 2017; 1(1): 34-43. doi: 10.18689/mjv-1000108
Copyright: © 2017 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
The human immunodeficiency virus (HIV) vaccine is urgently needed to curtail the global AIDS epidemic. Here, we report the findings from the Phase I clinical trial of a candidate multigene HIV-1 clade A DNA vaccine and demonstrate the induction of HIV1–specific immune responses.
Three groups of trial participants (21 healthy HIV-1-negative adult volunteers) were vaccinated four times with one of three different doses of DNA (0.25, 0.5 or 1.0 mg) administered intramuscularly. Vaccine-induced immune responses were measured by extensively validated methods, including ELISA, intracellular cytokine staining (ICS), IFNγ enzyme immunospot (ELISpot) assay and lymphocyte proliferation assay.
We found that our DNA vaccine was safe and well-tolerated at three used doses. Altogether, T-cell immune responses were elicited in all of 21 participants. We observed the increase in lymphocyte proliferation after fourth immunization that can show the advantage of fourfold against triple immunization. The frequency of detection positive cytokine responses decreases with increasing the vaccine dose. The humoral responses were induced in 5 people (24%). We didn't observe any correlation between the antibody production and the DNA-4 vaccine doses.
We also found the important correlation with our results obtained for the HIV specific immune responses in exposed seronegative individuals, i.e. TNFa production in immunized group.
Our findings of 100% immune reactivity of trial participants and the correlation of TNFa production with that in ESN individuals, may be promising indications for the possible efficacy of our candidate DNA vaccine.
Keywords: HIV-1; DNA vaccine; Clinical trial; Immune responses.
Introduction
The human immunodeficiency virus (HIV) vaccine is urgently needed to curtail the global AIDS epidemic. The results obtained in RV144 clinic trial of a canarypox vector prime/gp120 protein boost vaccine for HIV-1 suggest that neutralizing antibody responses can be partially protective against HIV-1 in low-risk heterosexual populations [1]. It is also known that T cells, particularly CD8+ T cells, exert some control over HIV-1 viremia and progression to disease in natural infection. In the acute stage of infection, decreased HIV-1 viremia is associated with the detection and expansion of virus specific CD8+ T cells prior to the detection of neutralizing antibodies [2-6]. That is why the induction of HIV-specific cell-mediated immunity may be necessary for preventive HIV vaccine, in combination with induction of neutralizing antibodies.is associated with the detection and expansion of virus specific CD8+ T cells prior to the detection of neutralizing antibodies [2-6]. In chronic HIV1 infection, about 5% of HIV infected patients do not progress to AIDS. Resistance to disease in these patients and resistance to infection in highly exposed seronegative patients is associated with different combinations of human leukocyte antigen (HLA) [7, 8] as well as with the detection of HIV-1-specific CD8+ T cells [9, 10]. It was demonstrated that in rhesus macaques a replicating cytomegalovirus vector expressing SIV antigens could eradicate early SIV infection 50% of SIVchallenged animals [11]. That is why the induction of HIVspecific cell-mediated immunity may be necessary for preventive HIV vaccine, in combination with induction of neutralizing antibodies.
Vaccines based on DNA technology represent a promising approach that has been shown to induce cell-mediated immune (CMI) responses [12-16].
Several veterinary DNA vaccines were licensed: one against the West Nile virus (WNV) in horses [17], one against infectious hematopoietic necrosis virus (IHNV) in salmon [18] and one against melanoma in dogs [19].
We developed candidate DNA vaccine expressing HIV-1 clade A Nef, Gag, RT and Env proteins. Preclinical studies demonstrated HIV-specific cell mediated immune responses induced in mice by the immunization with these DNA immunogens [20].
Here, we report the findings from the Phase I clinical trial of a candidate multigene HIV-1 clade A DNA vaccine and demonstrate the induction of HIV-1–specific immune responses.
Methods
Study vaccines
DNA-4 is a DNA vaccine developed by Biomedical Center (St. Petersburg, Russia) in collaboration with State Research Institute of Pure Biopreparations (St. Petersburg, Russia). It consists of four plasmids encoding consensus nef, gag, rt or gp140 sequences of HIV-1 FSU subtype A genes [20]. DNA-4 was manufactured by production facility of State Research Institute of Highly Pure Biopreparations (St. Petersburg, Russia) in accordance with the existing Russian federal regulations. The vaccine was formulated in a sterile saline solution with overall plasmid concentration of 1 mg/ml.
Phase I clinical study design and sample collection
The Phase I trial was conducted to access safety, tolerability and immunogenicity of the DNA-4 HIV candidate vaccine. The primary endpoints were the frequency of clinical adverse events and laboratory abnormalities, as well as the number of patients responded to injection of DNA-4 by cytokines expression and lymphocyte proliferation.
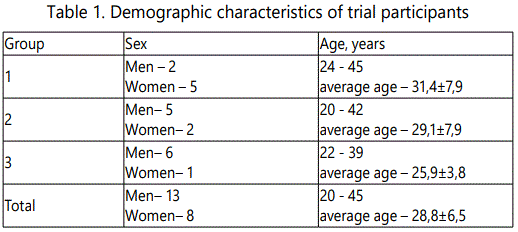
21 healthy HIV-1-negative adult volunteers aged 20−45 years of both genders were recruited. The individuals enrolled in this Phase I trial had no history of chronic, allergic and immunodeficient diseases, organ transplantions or psychiatric disorder, were negative in Hepatitis B and C viral tests. The pregnancy test for all female subjects was negative. All subjects were recruited at the single clinical trial site at the Pavlov State Medical University (St. Petersburg, Russia).
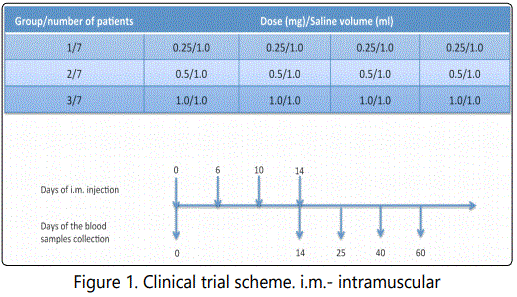
All trial participants were randomly divided into 3 groups. Each group of trial participants was vaccinated at four time points (days 0, 6, 10 and 14) with one of three different doses of DNA (0.25, 0.5 or 1.0 mg, starting with the least) administered intramuscularly into the right deltoid muscle (Figure 1). The use of each higher dose in the other group of participants was begun after obtaining the results, characterizing safety and low reactogenicity of a lower dose.
PBMC and plasma samples were collected at study days 0 (before immunization, for negative control), 14 (before immunizations), 25, 40, 60 to measure cell mediated (CM) and antibody immune responses (Figure 1).
Vaccine-induced immune responses were measured by extensively validated methods, including enzyme-linked immunosorbent assay (ELISA), intracellular cytokine staining (ICS), IFNγ enzyme immunospot (IFNγ-ELISpot) assay and lymphocyte proliferation assay (LPA) on cryopreserved peripheral blood mononuclear cells (PBMCs), as described below. Immune responses were assessed in different groups, at different visits and to different types of antigen.
Ethical compliance
The study was reviewed and approved by the Ethical Committee of the Ministry of Health of the Russian Federation (clinical trial approval number 283 from June 21, 2010, issued by the Federal Service for Surveillance in Healthcare of the Ministry of Health of the Russian Federation). Volunteers provided written informed consent following protocol review, and discussion and counseling with the clinical study team.
Peptide pools
A panel of 451 overlapping peptides spanning HIV-1 subtype A-Eastern European (EE) Env, Gag, RT, and Nef proteins was synthesized as previously described [21]. The peptides were 15 aa long overlapping by 11 aa and were represented in four different gene specific pools. Peptides were dissolved at a concentration of 55.6 mg/ml in 100% DMSO (Sigma-Aldrich). Pools of peptides were reconstituted in 90% PBS, 10% DMSO; the concentration of each individual peptide in the pools was 100 μg/ml. A cytomegalovirus, Epstein–Barr virus, and influenza virus (CEF) peptide pool (C.T.L.) was dissolved in DMSO and subsequently diluted 1:50 in 90% PBS, 10% DMSO to reach a final individual peptide concentration of 20 μg/ml.
IFNγ-ELISpot
The interferon IFNγ-ELISpot assay for HIV-1 peptidespecific T cells used the human IFNγ-ELISpot kit from Mabtech (Cincinnati, OH) with Millipore (Billerica, MA) IFNγ-ELISpot plates. The assay was performed as previously described [21]. Wells containing the medium without peptides served as the negative control and wells containing 1 μg/ml phytohemagglutinin (PHA) served as the positive control. A CEF peptide pool was used as the peptide positive control. Spots were counted and analyzed on a CTL immunospot reader and recorded as the mean spot-forming cells (SFC) per million PBMCs in triplicate wells. The final numbers of peptidespecific SFC were obtained by subtracting the background spots in the medium control wells.
A positive T cell response in IFNγ-ELISpot was defined as previously described [21]. Peptide pools with adjusted onesided p values of <0.05 were considered as positive. The mean background subtracted response for the peptide pool should be ≥50 SFU/106 PBMCs for the peptide pool to be considered positive.
Lymphocyte Proliferation Assay (LPA)
PBMC (1×106 /ml) were labeled with 5 mM carboxyfluorescein diacetate succinimidyl ester (CFSE; Sigma) for 8 min in the dark and then washed three times with PBS. Cells were then cultured in R10 medium at 37°C, 5% CO2 for 5 days with Nef, Gag, RT or Env peptide pools, R10 (negative control) or 5 μg/ml ConA (positive control). Cells were stained with CD8-PE-Cy5 (BD) and at least 100000 events were acquired using an EPICS XL (Beckman Coulter).
Using the results of the positive and negative controls the regions corresponding to non-dividing cells and the cells that divided one or two times were identified. Since for the vast majority of the patients the number of cell divisions in culture did not exceed two, for all trial participants only the percentage of the initial cells, dividing one or two times was determined. The number of PBMCs divided 0, 1 or 2 times was counted as N0, N1 and N2, respectively. For each trial participant the percentage of proliferating cells was calculated with respect to the corresponding arithmetic mean of the values of negative controls (N0к-, N1к- and N2к-, respectively). For each round of division the difference between the experiment and the mean control was calculated by the formula: Δι = Nι-(Nιk-×N0/N0k-), where ι is 1 or 2. The number of progenitor cells (K0), cells divided once (K1) and cells divided twice (K2) was found using formulas: K0 = N0 + K1/2, K1 = (Δ1 + K2/2)/2 and K2 = Δ2/4, respectively.
Final results were expressed with the following formula: Cμ= (Kμ/K0)×100%, where Cμ is the percentage of the initial cells dividing one or two times; and μ is 1 or 2.
Immune responses were considered as positive if:
1. The value of the immune response exceeds the upper limit of the 95% confidence interval of variation of the negative control.
2. The value of the immune response exceeds the values for the corresponding screening visit 1.
3. The value of an immune response of cells divided two times is valid, if it and the corresponding percentage of cells divided one time satisfies the criteria 1 and 2 simultaneously.
Intracellular Cytokine Staining (ICS)
Production of the intracellular IFNγ, TNFa, and IL-2 by CD3+CD8+ and CD3+CD8- cells was examined in response to HIV peptide pools using multiparameter flow cytometry. According to our preliminary research (data not shown) the CD3+CD8- cells correspond toCD3+CD4+ significantly, so we measured helper T cells as cells with CD3+CD8- phenotype. The assay was performed as previously described [21].
Cells incubated with costimulatory antibodies only were included in every experiment, to control for spontaneous production of cytokines and activation of cells before the addition of peptides. Cells stimulated with0.025 μg/ml Phorbol 1,2-myristate 1,3-acetate (PMA, Sigma) were used as a positive control for lymphocyte activation.
Flow cytometric analysis was performed on an EPICS XL (Beckman Coulter). At least 50 000 events, gated on small lymphocytes, were acquired and analyzed using WinList 32 (Software House Inc., USA). The final numbers of cells expressed cytokines in response to specific peptide stimulation were obtained by subtracting the background cell number in the medium control tubes.
A positive T cell response in ICS was defined as previously described [21]. A positive T cell response required the acquisition of 50,000 CD3+ lymphocytes, and the frequency of cytokine-producing T cells in antigen-stimulated cells had to be at least two times that of nonstimulated cells.
ELISA
ELISA was used to detect the gp120, Nef, RT, P17 or P24 specific IgG responses. These recombinant proteins were prepared as previously described [22; 23]. 96-wells plates were coated with antigens in PBS at 37°C for 1 hour; plates were then washed and blocked in blocking buffer (PBS, 0.05% Tween 20, 2% skim milk) for 30 minutes at 37°C. Serum dilutions 1:100 were prepared in blocking solution and incubated in the plates for 1 hour at 37°C. Conjugate was diluted to 1:2000 in blocking buffer and then incubated on the plates for 1 hour at 37°C. Substrate-reagent solution was added to the plates and incubated for 0.5 hour at 37°C. Between steps, the plates were washed 5 times with PBS, 0.05% Tween 20. Reaction was stopped with sulfuric acid. Assays were read immediately at 450 nm using the iMark (Bio-Rad).
Statistical analysis
Microsoft Office Excel 2003, Prism 4.03 and the opportunities provided by online resource were used for statistical analysis of the research results.
Group comparisons were made by Fisher Exact Probability Test, Chi-Square Test of Association, Mann-Whitney U-test and Kruskal-Wallis test followed by pair wise comparisons using Dunn's Multiple Comparison Test (ANOVA).
Results
Adherence and tolerability
21 healthy HIV-1-negative adult volunteers aged 20−45 years of both genders were recruited (Table 1).
Vaccinations were completed in 20 (95%) of 21 subjects, but the data from all 21 subjects were included in the analysis. One of the trial participants from group 2 became sick, for reasons not related to the application of the vaccine. This patient was vaccinated only two times, but all blood samples at all timepoints were collected and analyzed. All subjects were monitored for clinical adverse events, reactogenicity, laboratory abnormalities, pregnancy, and HIV infection through 60 days of follow-up.
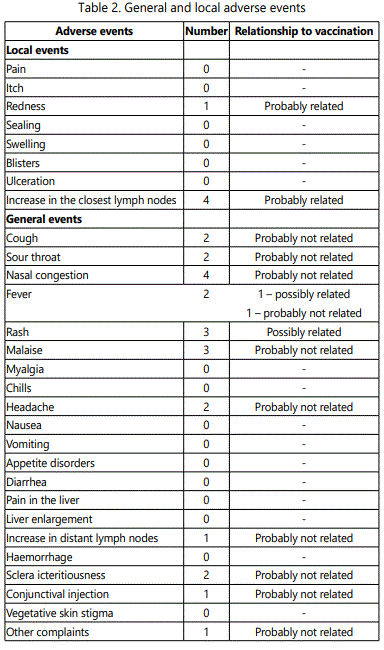
Overall the DNA-4 vaccine was well tolerated with no dose-related toxicities. There were no episodes of severe reactogenicity. The most frequent adverse events were skin reactions and increase in axillary lymph nodes (Table 2).
Cell mediated immune responses
Cellular immune responses induced in patients after immunizations were measured using ICS, IFNγ-ELISpot and LPA. Using these methods immune responses were analyzed in all 21 trial participants (Figure 1).
Results of positive controls for all tests used in this study were adequate and relevant (data not shown).
Immune responses at different vaccine doses
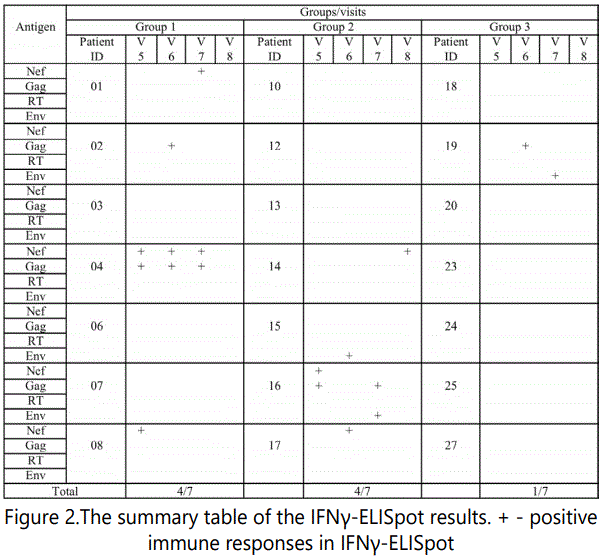
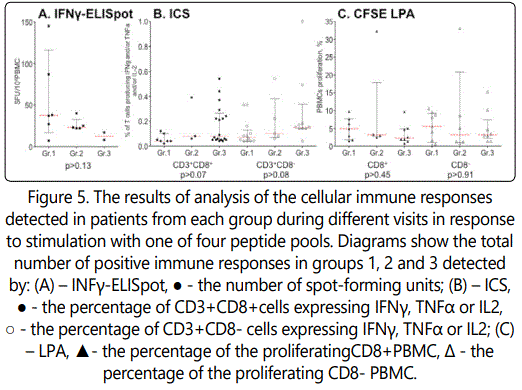
Cellular immune responses were detected in patients from different groups immunized with different vaccine doses in response to stimulation with Env, Gag, RT and Nef antigens. Four subjects from group 1 (4/7) and four subjects from group 2 (4/7) and one subject from group 3 (1/7) had positive IFNγ-ELISpot immune responses (Figure 2).
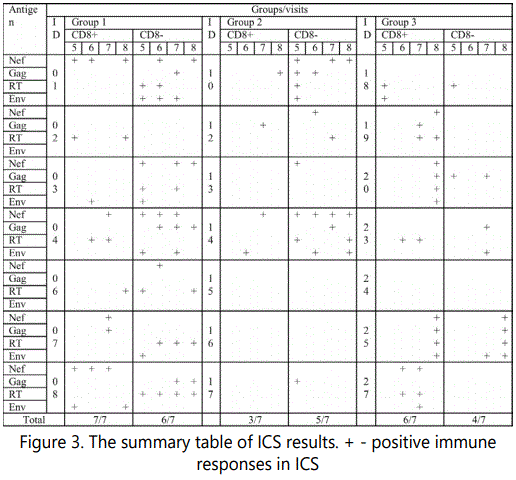
The production of cytokines IFNγ, IL2, and TNFα by CD3+CD8+ and CD3+CD8- cells was measured using flow cytometry. Specific cytotoxic and helper immune responses were detected in all vaccine groups: 7 persons (7/7) in group 1, 5 persons (5/7) in group 2 and 6 persons in group 3 (6/7) (Figure 3).
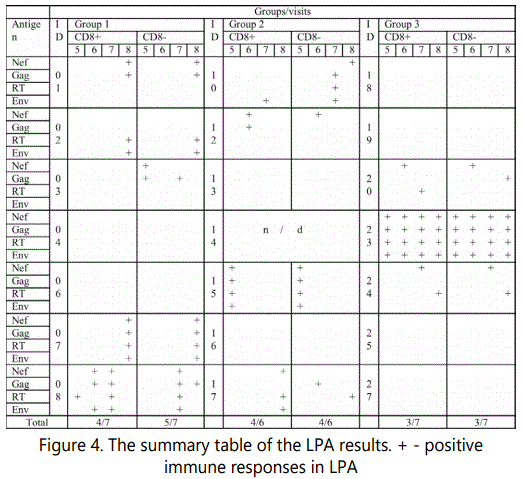
Positive lymphocytes proliferation (both in CD8+ and CD8-) was detected in five persons from group 1 (5/7), in four persons from group 2 (4/6), and in three persons from group 3 (3/7) (Figure 4).
Differences between these groups were statistically insignificant (p>0.15 for ELISpot; p>0.74 for ICS; p>0.62 for LPA; Fisher Exact Probability Test).
Quantitative data for positive immune responses in different groups are presented in the Figure 5. Every dot on the picture presents the number of spot-forming units (for IFNγ-ELISpot); the percentage of cells expressing IFN γ, TNF α or IL2 (for ICS); or the percentage of proliferating PBMC (for LPA) detected in patients from each group during different visits in response to stimulation with one of four peptide pools. Analysis using Kruskal-Wallis test showed no significant difference between the groups of trial participants in any of the assay, indicating that all three doses had the same effect (Figure 5).
Positive cytokine responses were detected in 52 (23%) of 224 experiments (5-8 visits) in group 1, in 24 (11%) cases in group 2 and in 31 (14%) cases in group 3. So frequency of detection positive cytokine responses decreases with increasing vaccine dose (c2= 14.16, p<0.0009 in comparison of groups 1 and 2; c2= 5.92, p<0.015 in comparison of groups 1 and 3).
Sixteen of 21 persons had CD3+CD8+ immune responses and 15 persons had CD3+CD8- responses (Figure 3). Intergroup difference between cytotoxic and helper immune responses was not observed (p>0.73Fisher Exact Probability Test).
We analyzed mono- and poly- cytokine production in response to specific antigen stimulation. Expression of one of cytokines (IFNγ, TNFα or IL2) was detected in 7 persons (7/7) from group 1, in 3 persons (3/7) from group 2 and in 6 persons (6/7) from group 3. Seven patients from group 1 (7/7), 5 from group 2 (5/7) and 6 from group 3 (6/7) had production of two cytokines. Six subjects from group 1 (6/7), 5 subjects from group 2 (5/7) and 2 subjects from group 3 (2/7) responded to antigen stimulation by expression of all three cytokines simultaneously (Table 3). The differences between groups were insignificant (p>0.66, Fisher Exact Probability Test).
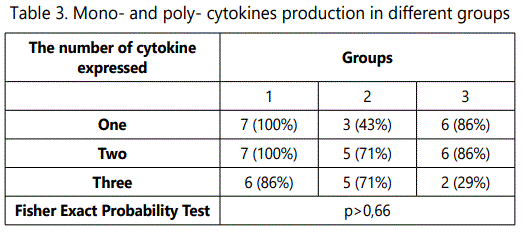
We conducted 112 specific tests for cytokines expression in CD3+CD8+ and CD3+CD8- lymphocytes in each group (7 participants in group, 4 visits, 4 antigen stimulations). Table 3 presents the number of positive cases in these experiments. It should be noted that with increasing doses of DNA (0.25 mg to 1 mg) there was an increase in the frequency of CD3+CD8+ monospecific immune responses but reduction of CD3+CD8+ polyspecific (two or three cytokines simultaneously) immune responses. Furthermore the expression of several cytokines simultaneously by CD3+CD8- lymphocytes decreased with increasing doses of DNA vaccine (Table 4).
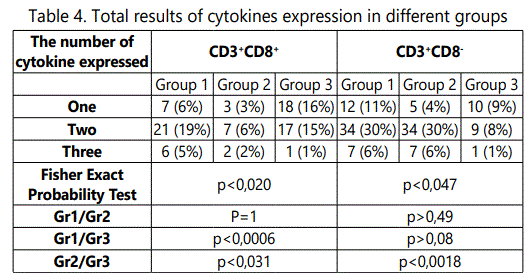
We observed also that the increasing doses of the DNA vaccine correlated with significant increase in the number of CD3+CD8+ lymphocytes expressing IL-2, and the number of CD3+CD8- lymphocytes expressing TNFα. On the other hand, there is a significant drop in the number of CD3+CD8- lymphocytes responsible for simultaneous expression of IFNγ and TNFα.
Immune responses at different visits
Immune responses were measured at different visits starting from the fifth (after the third vaccination) till eighth (60th day of follow up).
After three immunizations (visit 5) positive IFNγ-ELISpot immune responses were detected in 3/21 patients and after four immunizations (visits 6-8) – in 8/21 patients (Figure 2). There was no significant difference between visits in these data (p>0.15, Fisher Exact Probability Test).
The overall cytokine expression at 5th visits was observed in 13/21 subjects, at 6th visit – 11/21 subjects, at 7th visit – 13/21 and at 8th visit – 12/21 (Figure 3). Like in the case of IFNγ-ELISpot there were no statistically significant differences between different visits (p>0.80, Fisher Exact Probability Test).
Only four subjects (4/20) demonstrated lymphocyte proliferation after three vaccinations (visit 5; both for CD8+ and CD8- cells). After four immunizations (visits 6, 7, 8) proliferation was detected in eleven individuals (11/20) (Figure 4). The increase in lymphocyte proliferation after fourth immunization is statistically valid (p<0.019, Fisher Exact Probability Test) and shows the advantage of fourfold against the triple immunization.
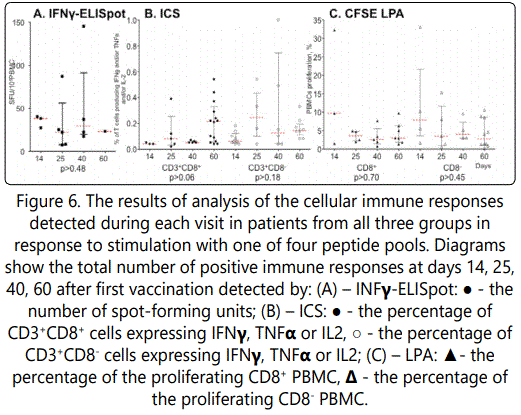
Quantitative data for positive immune responses at different visits are presented in the Figure 6. Every dot on the picture presents the number of spot-forming units (for IFNγELISpot); the percentage of cells expressing IFNγ, TNFα or IL2 (for ICS); or the percentage of proliferating PBMCs (for LPA) detected in patients from all three groups at each visit in response to stimulation with one of four peptide pools. Analysis using Kruskal-Wallis test showed no significant difference between visits of trial participants in any of the assay, indicating that at all four time points we have the same effect (Figure 6).
Dependence of the immune responses on the type of antigen
Immune responses to four antigens used in the vaccine (nef, gag, rt and gp140) were measured. Six of the responders (6/21) were IFNγ-ELISpot positive for Nef peptide pool stimulation, four (4/21) - for Gag and three – for Gp140. Immune responses to RT peptide pool were not detected (Figure 2). Differences between immune responses were insignificant (p>0.60, Fisher Exact Probability Test).
According to ICS results, 14 patients (14/21) produced cytokines in response to stimulation with Nef, 12 (12/21) in response to Gag, 12 (12/21) in response to Gp140 and 16 (16/21) in response to RT (Figure 3). Nine (9/21), 6 (6/21), 8 (8/21) and 7 (7/21) subjects demonstrated cytokine expression by CD3+CD8+ PBMC in response to Nef, Gag, RT or Gp140 peptide stimulation, respectively; CD3+CD8- PBMC expressed cytokine in response to Nef, Gag, RT or Gp140 stimulation in 9 (9/21), 9 (9/21), 10 (10/21) and 8 (8/21) subjects, respectively (Figure 3). Differences between all these data were statistically insignificant (p>0.54; p>0.84; p>0.93, respectively. Fisher Exact Probability Test).
Lymphocyte proliferation was observed in response to stimulation with all peptide pools: in 11 persons (11/20) - to Nef or Gag, in 9 persons (9/20) – to RT, and in 7 persons (7/20) – to Gp140 (Figure 4). There were no significant differences between these results (p>0.74, Fisher Exact Probability Test).
Quantitative data for positive immune responses to different antigens are presented in the Figure 7. Every dot on the picture presents the number of spot-forming units (for IFNγ-ELISpot); the percentage of cells expressing IFNγ, TNFαor IL2 (for ICS); or the percentage of proliferating PBMCs (for LPA) detected in response to stimulation with one of four peptide pools during all visit in patients from all three groups, respectively. Analysis using Kruskal-Wallis test showed no significant difference between different antigens in any of the assay, indicating that each of four antigens has similar effect. (Figure 7).
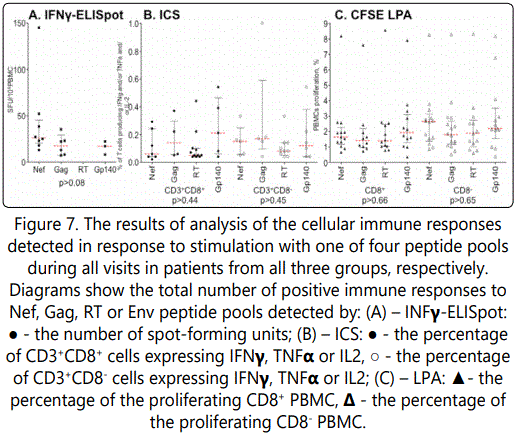
Figure 8 shows the overall results for CD3+CD8+ and CD3+CD8- lymphocytes from participants expressing one, two or three cytokines simultaneously.
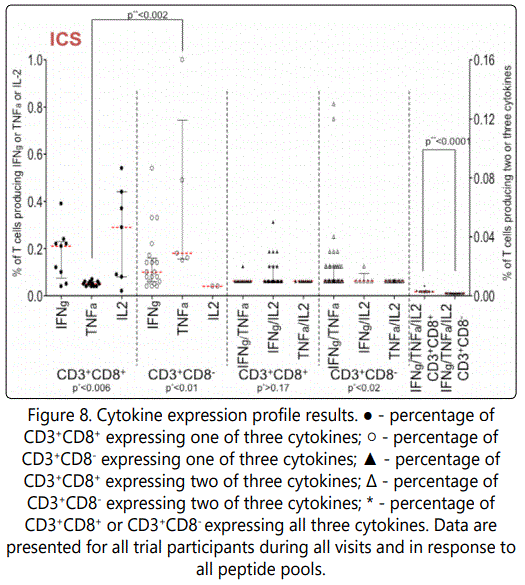
We noted statistically valid differences between T cells proportions of T cells demonstrating monospecific immune responses (р<0.006 for CD3+CD8+ and р<0.01 for CD3+CD8- , Kruskal-Wallis test); as well as between helper proportions of helper lymphocytes expressing different combination of two cytokines (р<0.02, Kruskal-Wallis test) (Figure 8).
Statistically meaningful differences between CD3+CD8+ and CD3+CD8- cells in expression of TNFα or IFNγ/TNFα/IL2 positive lymphocytes were found (p<0.002 and p<0.0001, respectively. Mann-Whitney U-test) (Figure 8).
Vaccine specific antibody responsesp Vaccine specific antibody responses were observed by ELISA in postimmunization serum of 5 trials participants: 2 (2/7) subjects in group 1, 2 (2/7) subjects in group 3, and 1 (1/7) subject in group 2. So the frequency of antibody responses was independent of the dose.
Of five subjects two had detectable antibody against Nef, one had detectable antibody against P17 and one had detectable antibody against Gp120. In all cases, the antibody titer did not exceed 100 and grew until the last visit.
Discussion
The phase I trial was conducted to access safety, tolerability and immunogenicity of the DNA-4 HIV candidate vaccine. DNA-4 was safe and well-tolerated at three used doses, with the majority of subjects remaining free of local and general symptoms (Table 2).
Immune response induced by the DNA-4 vaccine was assessed using ELISA, IFNγ-ELISpot, ICS and LPA. Our results demonstrate that our candidate DNA vaccine is immunogenic: 9 patients were IFNγ-ELISpot reactive, 18 patients expressed cytokines to specific antigen stimulation, and 12 patients had positive lymphocyte proliferation. For some patients immune responses were detected using several methods (Table 5).
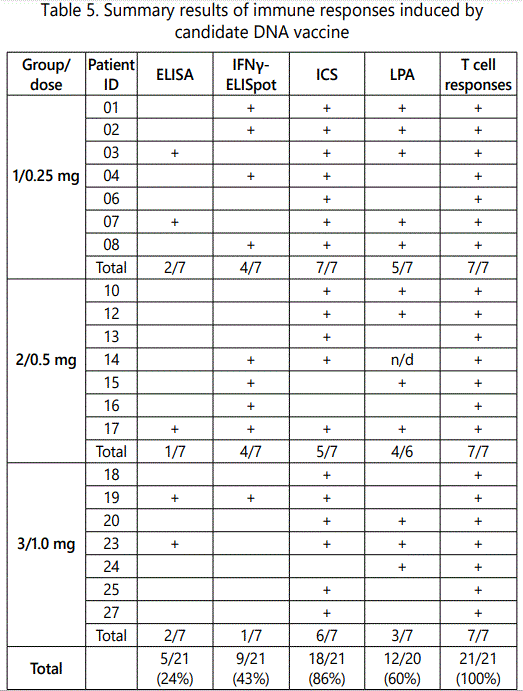
Even the patient #12, who receive only 2 injections of the DNA-4 vaccine (the dose 0.5 mg) and left the study because of sickness not related to the vaccination (cold), demonstrated the induction of cytotoxic as well as helper lymphocytes proliferating after the antigen stimulation.
Altogether, T-cell immune responses were elicited in all of 21 participants (Table 5). The data obtained by other authors who used different candidate HIV vaccines, to our knowledge, never demonstrated the 100% immune reactivity. Thus, in ACTG 5176 trial only 11 of 16 treated HIV-infected adults (68.8%) developed low-magnitude T-cell responses to Pol or Gag peptides [15]. In HVTN 502/Merck 023 trial (STEP Trial) positive IFNγ-ELISpot results were available for 217 of 257 study participants (84%) at week 30 [24]. But in these studies only IFNγ-ELISpot characteristics have been analyzed, while in our study the 100 % responsiveness is calculated as combined responsiveness to all immunogens, all doses, all time points, and all methods.
All three doses had the same effect on immune responses in trial participants. The frequency of detection positive cytokine responses was the only parameter that decreased with increasing the vaccine dose (c2= 14,16, p<0,0009 in comparison of groups 1 and 2; c2= 5,92, p<<0,015 in comparison of groups 1 and 3). Graham and co-authors demonstrated a trend toward a grater magnitude and frequency of T cell responses in recipients of 4- or 8-mg dose of multiclade HIV-1 DNA candidate vaccine than in recipients of the 2-mg dose, but that difference was not statistically significant [13].
Other authors showed in macaques that increasing amounts of env DNA resulted in greatly enhanced Env antibody titers without significantly affecting the levels of anti-Env cellular immune responses [25]. They also found that a balanced ratio of Gag and Env was important to avoid immunological interference and to achieve both maximal humoral responses against Env to prevent virus acquisition and maximal cytotoxic T cell responses against Gag to prevent virus spread [25].
We observed the increase in lymphocyte proliferation after fourth immunization that can show the advantage of fourfold against triple immunization (p<0.019).
The humoral response was induced in 5 people (24%). The titer of HIV-specific antibodies didn't exceed 1/100. This correlates with the results of our preclinical studies, where the induction of predominantly cellular immune response after vaccination of mice with DNA vaccine was observed [20], and also with the results of others obtained in humans [14].
We didn't observe any correlation between the antibody production and the DNA-4 vaccine doses. At the same time Graham and co-authors demonstrated the correlation of the frequency of antibody responses in human with the dose of the DNA vaccine. But it should be noted, that the doses used in that study (2-8 mg) exceeded our doses, and the authors didn't detect antibodies after injection of the least dose (2 mg) [13].
The immune responses detected in trial participants were weak that agrees with the literature data. DNA vaccines are considered to have a low immunogenicity and in the majority of cases require additional booster for induction of the stronger immune response. Thus, Mullligan and co-authors didn't observe immune responses after double injection of naked plasmid DNAs encoding Gag, Pol, Env, Tat, Rev, Vpu genes (0,3 and 3 mg) in humans [26].
There are several approaches to enhance immune responses induced by DNA vaccination. One of them is to use different prime-boost combination. Recombinant MVA [26], recombinant Ad5 [27; 28], recombinant vaccinia virus [29] and proteins [30] have been used as components of prime-boost vaccination. For example Shneider and co-authors demonstrated that primeboost vaccination with a combination of proteosome-degradable and wild-type forms of two influenza proteins leads to augmented CTL response [31].
Adjuvants [32] and different delivery methods of plasmid DNA are also used to enhance the immunogenicity of DNA vaccines. For example, Vasan and co-authors demonstrated significant increase of the immunogenicity of the HIV candidate DNA vaccine ADVAX when an electroporation in vivo was applied [33].
Another interesting approach for enhancing the T-cell immune response induced by the DNA vaccine was implemented by Yan and co-authors in cynomolgus macaques. Animals were immunized I.M. divided into two doses administered on consecutive days (Study Days 1, 2; 29, 30; and 57, 58). Each immunization was given in 2 mL on each day (total of 3 mg/day) into four sites (0.5 mL/site). In this way T-cell immunity in four independent lymph nodes was stimulated. Immune responses were measured by the IFNγ-ELISpotand LPA. It was shown that such vaccination with SIV DNA immunogens could induce SIV antigen-specific CD8+ T cells that have high proliferative capacity [34].
Not only the magnitude of immune responses, but also the character of immunity may be a main factor for achieving protective effect. The combination of T-cell and humoral immune responses is considered to be necessary for prevention of the HIV infection. It was demonstrated in the Step Study of MRKAd5 HIV-1 gag/pol/nef vaccine that the vaccine elicited IFNγ-ELISpot responses in 75% of the 25% random sample of all vaccine recipients. But the study was stopped for futility; there was an increase the number of HIV-1 infection in male vaccine recipients [35]. It was also found that in general the total expression of IFNγ, CD107a, or MIP-1β correlated with viral inhibition. Likewise, TNFa or IL-2 secretion was not observed to be characteristic of HIV-inhibitory CD8 T-cells [36].
In total a lot of different immune correlates, including exotic parameters, are used for assessment vaccine efficacy, but specific correlates of protection for vaccines against HIV remain unclear because of absence of successful trials and adequate animal model.
In our study despite the limited magnitude of the immune response we found the important correlation with our results obtained for the HIV specific immune responses in exposed seronegative individuals (ESN). Thus, in our previous study the increase of TNFa expression by the CD4+ T cells after specific peptide stimulation was demonstrated for ESN individuals with high risk of HIV infection [21]. In trial participants after immunization with candidate DNA vaccine we observed the similar effect (Figure 8). Such correlation could be an indication on the possible effectiveness of our DNA vaccine.
To date the RV144 study conducted in Thailand, with a canarypox-vectored prime followed by a protein vaccine boost with the gp120, is the only HIV vaccine study to show some protection (vaccine efficacy 31.2%) against HIV infection. The study of immune correlates resulted in the hypotheses that V1V2 antibodies may provide the protection against HIV-1 infection, whereas high levels of Env-specific IgA antibodies may reduce the effects of protective antibodies [37].
For protection of horses against West-Nile virus the single immunization with 1 mg of DNA vaccine is effective. All vaccinated horses remained healthy after virus challenge [38].
So, even concentrations of DNA that demonstrate weak immune responses or do not demonstrate the induction of immune correlates measured by available methods may be effective as vaccines. Our finding of 100% immune reactivity of trial participants, although at different conditions in different patients, as well as the correlation of TNFa production with that in ESN individuals, may be indications for the possible efficacy of our candidate DNA vaccine.
Acknowledgments
This work was supported by Russian HIV Vaccine Program. A.P.K., E.A., S.V., A.M. are currently supported by Russian Science Foundation grant # 15-14-0026 to A.P.K.
Conflict of interest
The authors declare that they have no Conflict of interest.
References