Research Article
Determination of HCV Genotypes and Viral Loads in Chronic Hepatic Sudanese infected Patients
Department of Microbiology, Medical Laboratories Sciences, Omdurman Islamic University, Omdurman, Sudan
*Corresponding author: Mohammed EH Our Nasseir, Faculty Department of Microbiology, Omdurman Islamic University, Omdurman, Sudan, Tel: +249912959304, E-mail: ornaser@hotmail.com
Received: April 23, 2019 Accepted: May 3, 2019 Published: May 10, 2019
Citation: Nasseir MEH, Elawad HE, Osman NAM. Determination of HCV Genotypes and Viral Loads in Chronic Hepatic Sudanese infected Patients. Int J Microbiol Curr Res. 2019; 1(1): 29-32. doi: 10.18689/ijmr-1000106
Copyright: © 2019 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Background: Knowledge of Hepatitis C virus (HCV) genotypes is significant for arranging treatment regimes. Quantitative HCV RNA testing provides prognostic data useful in monitoring the efficacy of antiviral therapy.
Methods: A total of 1203 serum samples were collected from individuals attending outpatients units at Khartoum State and Gezira State. The study population comprises two groups. Blood donors study groups (n=600) and chronic hepatic patients during the course of HCV infection (n=603). Serum samples was screened using enzyme linked immune-sorbent assay (ELISA) (Biokit, A.S. Spain®). HCV positive samples (n=100) were quantified by HCV Real-TM Quant SC (Sacace Biotechnologies Italy®).
Results: Hundred HCV sero-positive samples were subjected to genotyping and quantitative analysis of these samples using RT-PCR, HCV genotype 4 was the predominant genotype (92%) followed by genotype 2 (4%), genotype 1 (2%) and 3 (2%) in different groups. The average viral load of the patients infected with genotype 4 was higher than an average viral load of the patients infected with genotypes 1,2 and 3.
Conclusions: The present study highlighted that genotype 4 is the predominant genotype in Sudanese hepatic patients followed by genotype 2. The severity of liver disease was more among genotype 4 patients as assessed by a higher viral load.
Keywords: Hepatitis C virus; Genotypes; Viral loads; Chronic hepatic.
Introduction
Hepatitis C infection (HCV) was recognized in 1989 and was observed to be in charge of 70-90% of post-transfusion hepatitis in all countries wherever blood was tested for Hepatitis B infection (HBV) [1]. Atleast six major HCV genotypes involving various, more closely related subtypes have been identified [2]. HCV genotypes show significant variations in their worldwide distribution and predominance, making genotyping a helpful technique for determining the source of HCV transmission in an infected localized population [3]. Infection by HCV is the main cause of chronic liver disease worldwide [4], the patients uninformed of their disease and at risk for developing cirrhosis and hepatocellular carcinoma [5]. HCV is an enveloped virus that has a place with the family Flaviviridae. Its linear positive-stranded RNA genome of approximately 9.6 kb in length encodes both structural (E1, E2 and core) and nonstructural (N2, NS3, NS4a/b, p7 and NS5a/b) proteins in a single open reading frame, flanked by short conserved untranslated regions (UTRs) located at the 5\ and 3\ ends of the genome that are required for viral replication and protein translation [6,7]. Humans appear to be the only source of infection and inoculation with blood and blood products is that the most recognized mode of transmission [8]. The majority of the acute infections are symptomless with around 10% of patients having a mild flu-like illness with jaundice and an increase in blood serum aminotransferase. Hepatitis C virus (HCV) is the dominant part of parenteral and sporadic non-A-non-B hepatitis cases. At least 70% of infected patients develop chronic infection and approximately 20% progress to cirrhosis of the liver [9-11]. In spite of the fact that hepatitis C virus (HCV) infection is a major cause of chronic liver disease around the world, the virus has not yet been cultured in vitro and little is understood regarding its biological and physicochemical properties. Till sensitive and accurate test become available, diagnosing of HCV is usually done by exclusion of a high-risk individual with negative markers for HAV and HBV and also on clinical and epidemiologic features of the individual patients [12,13].
HCV RNA can be detected fourteen days after disease and anti-HCV antibodies are typically positive six weeks from infection [14]. Sensitive methods or tests also are currently available for the detection of viral nucleic acid antigens from nucleocapsid regions have been used to develop ELISA, the present assay ELISA-3 (3rd generation ELISA) incorporate nonstructural recombinant antigens three, four and five (NS3, NS4, and NS5) regions. The patients infected with HCV develop antibodies to numerous structural and nonstructural viral proteins. Sensitive methods or tests also are currently obtainable for the detection of viral nucleic acid. For example RT-PCR can detect HCV-RNA in the blood, however it is possibly used when serological tests gave obscure results as were as and in selecting for, and estimating response to therapy [15,16]. The absence of convenient culture system for HCV implies the utilization of molecular biology to evaluate viremia. Direct hybridization of serum samples is possible, however is hampered by the low virus concentration in many patients. Amplification of viral nucleic acid by the polymerase chain reaction (PCR) gives an exceedingly sensitive and antigen-antibody-independent technique to identify continuous viral infection. Although a positive PCR assay is not absolute verification of HCV viremia, it strongly suggests active virus production inside the body [17]. The treatment plan (combination of compounds, dosages, and duration) and therefore the virological follow-up for management of antiviral treatment in chronic HCV hepatic patients is a very important, but to guarantee good monitoring of the treated patients, doctors require fast, reproducible, and sensitive molecular tools with a large scope of detection and quantification of HCV RNA in blood tests.
Objectives
In the current study, we aimed to quantify and genotyping HCV in patients suffering from chronic hepatitis during the course of infection.
Materials and Methods
Enrollment of patients
Serum samples were obtained from subjects attending out-patients units in Khartoum State and Gezira State medical centers.
Sample collection
Blood samples were taken from 1203 individuals, including chronic hepatic patients and normal healthy blood donor upon their consent. All serum was separated from whole blood within 6 h of collection, and stored at -20°C until testing.
Extraction of HCV RNA
Viral RNA was extracted from serum samples with QIA amp viral RNA mini kit (Qiagen, Hilden, Germany®) following the manufacturerʼs directions wherever the input volume of blood serum was 170 μL and therefore the output was 60 μL.
Reverse transcription of HCV RNA
Reverse transcription was carried out using RT-G-mix-1 and RT-mix., were thawed, vortex and centrifuged briefly. Reaction mix was prepared: by added 5.0 μl RT-G-mix-1 into the tube containing RT-mix and vortex for at least 5-10 seconds, centrifuged briefly. 6 μl M-MLV were added into the tube with reagent. Then it were mixed by pipetting, vortexes for 3 sec, centrifuged for 5-7 sec (must be used immediately after the preparation). 10 μ of reaction mix were added into each sample tube. 10 μl RNA samples were pipetted to the appropriate tube. A re-centrifuge all the tubes with extracted RNA for 2 minutes were done at maximum speed (12000-16000 g) and carefully supernatant were taken. Tubes were placed into thermal cycler and incubated at 37°C for 30 minutes. Each obtained cDNA sample was diluted 1:2 with TE-buffer and centrifuged briefly the tubes. cDNA specimens could be stored at -20°C for a week or at -70°C up to one year.
Results
Analytical specificity and linearity
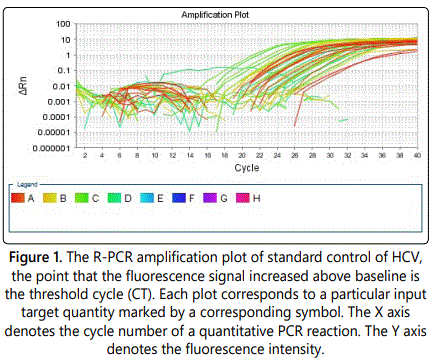
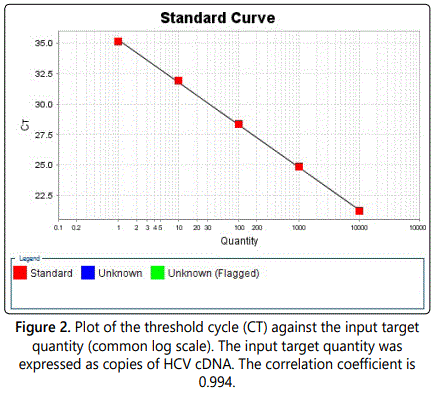
The standard calibration curve was generated by using the Smart Cycler II software and serial dilution. The analytical specificity of the primers and the probes was validated with 80 negative samples. They did not generate any signal with the specific HCV primers and probes. The specificity of the kit HCV Real-TM Quant was 100%. The linearity of the HCV Real-TM Quant assay was tested with the HCV RNA Standard and its dilution using HCV-negative human plasma. Each dilution was analyzed three times and the mean HCV RNA titer of each sample was determined (Figure 1).
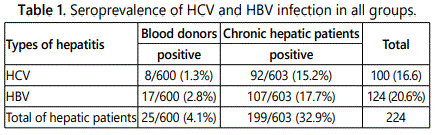
A total of 1203 samples were obtained from chronic hepatic patients during the course of infection as study groups (n=600) and normal healthy blood donors represent a control (n=603). The collected samples were screened for the HCV and HBV antibodies using ELISA. Of the 600 normal healthy blood donors which were screened for the presence of anti HCV/HBV antibodies, 8 out 600 (1.3%) showed HCV positive while 17 out of 600 (2.8%) were HBV positive. Serological screening carried out for chronic hepatic patients during the course of infection. We found that 92 out 603 (15.2%) were positive for HCV antibodies, while 107 out of 603 (17.7%) showed positive HBV antibodies (Table 1).
Detection of Plasma cell-free HCVRNA in chronic hepatic patients
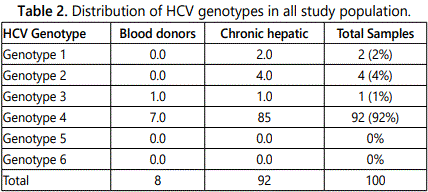
The HCV antibody positive samples were tested for the presence of HCV RNA. A molecular study was conducted to investigate the prevalence of Hepatitis C virus genotypes in HCV infected population of Khartoum state. 100 HCV seropositive samples were subjected to genotyping and quantitative analysis of these samples using RT-PCR, HCV genotype 4 was the predominant genotype (92%) followed by genotype 2 (4%), genotype 1 (2%) and 3 (2%) (Table 2).
Quantitative HCV RNA-specific PCR
Viral load quantification was carried out by Taqman real time PCR system in all 100 HCV RNA positive patients and was compared between the four groups of genotypes. The average viral load of the patients infected with genotype 4 was higher than average viral load of the patients infected with genotypes 1.2 and 3 (2.76 × 103–9.3 × 106) copies/ml.
Discussion and Conclusion
In the present study the seroprevalence, genotyping and estimation of viral load (viremia) of viral hepatitis C was evaluated in sera of different groups, include blood donors and hemodialysis patients, randomly selected in Khartoum area and screened by ELISA (third generation) using recombinant HCV-antigens, as well as real-time polymerase chain reaction (RT-PCR) for genotyping and quantitative viral load assay were estimated in positive individuals for HCV. In blood donors study population of 8/17/600 about 1.3/2.8%, samples showed positive results of HCV/HBV-antibodies respectively, when tested by ELISA using recombinant HCV-antigens. A similar study was previously done in Juba in Southern Sudan and 3% of the studied populations were found positive for HCV- antibodies [18]. Similarly, El-Hazmi M [19] also reported or highlights the prevalence rates of HBV and HCV (1.5% and 0.4%) respectively among completely different groups. The prevalence differs from one group to another, being the most reduced among Saudi and young donors. Accordingly, extensive recruitment of Saudi and young donors should help ensure a long-term increase in the blood supply without risk. Another investigation concluded that the prevalence of HCV infection in the population recruited from different health centers in Jordan is comparatively low and estimates a prevalence of 0.42% among all age groups and 0.56% among those aged >15 years old [20,21]. Low values of both this study and others suggest that HCV infection is not an endemic disease in healthy blood donors. In this study HCV infection has been found high among hemodialysis patients (15.2%) of the whole hemodialysis samples (92/603), have been found positive to HCV-antibodies, when tested by ELISA using recombinant HCV-antigens, whereas 107 (17.7%) of HBV positive antibodies were found when tested using same technique. This is in agreement with Suliman SM et al. [22]. Similarly Sato S et al. and Taura Y et al. [23,24] also reported that the prevalence of HCV infection among dialysis patients is generally matched higher than that among healthy blood donors. Dialysis patients have an increased risk of exposure to parenterally transmit hepatitis virus. Hemodialysis machine may represent a hazard of HCV transmission of HCV antibodies positive patients. The present study is in support of the positive, have a history of presentation to a dialysis machine. Once more results are in agreement with those obtained by Pereira BJ et al. [25]. However, the dialysis process itself and also the level of a hygienic standard may influence the risk level of HCV infection. Another study in Saudi was reported that the overall of anti-HCV antibodies were detected in 7.3% (1124/15323) of the studied individuals [26]. Abdel-Aziz F et al. reported that the high rate of anti-HCV prevalence has been assessed in Egypt of the total samples 24.3% (973/3,999) that showed the highest report in a community-based study through all age groups as well as reflected that HCV was endemic in Egypt [27]. In conclusion, the rate of risk factors that might contribute to HCV infection, we found patients had chronic disease in about 92%, this might represent the probability of hemodialysis machine in the transmission of the disease. While patients have received a blood transfusion and who have a history of surgery represented 24% and 6% respectively. Tattooing 3% and alcohol take 0% have no role in predisposition to the disease.
References
- Kuo G, Choo QL, Alter HJ, et al. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989; 244(4902): 362-364.
- Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000; 13(2): 223-235.
- Larke B, Hu YW, Krajden M, et al. Acute nosocomial HCV infection detected by NAT of a regular blood donor. Transfusion. 2002; 42(6): 759-765.
- Pearlman BL. Hepatitis C infection: a clinical review. South Med J. 2004; 97(4): 364-373.
- Sarbah SA, Gramlich T, Younoszai A, et al. Risk factors for hepatocellular carcinoma in patients with cirrhosis. Dig Dis Sci. 2004; 49(5): 850-853.
- Bradley JS, Graham S, Picchio GR, Vugia DJ, Kharrazi M. Prevalence of hepatitis C virus antibody in newborn infants in southern California in 2003. Pediatr Infect Dis J. 2011; 30(7): 618-620.
- Bradley DW. Studies of non-A, non-B hepatitis and characterization of the hepatitis C virus in chimpanzees. Curr Top Microbiol Immunol. 2000; 242: 1-23.
- Aach RD, Yomtovian RA, Hack M. Neonatal and pediatric posttransfusion hepatitis C: a look back and a look forward. Pediatrics. 2000; 105: 836-842.
- Daniel HD, Grant PR, Garson JA, Tedder RS, Chandy GM, Abraham P. Quantitation of hepatitis C virus using an in-house real-time reverse transcriptase polymerase chain reaction in plasma samples. Diagn Microbiol Infect Dis. 2008; 61(4): 415-420. doi: 10.1016/j.diagmicrobio.2008.04.001
- Smith DB, Lawlor E, Power J, et al. A second outbreak of hepatitis C virus infection from anti-D immunoglobulin in Ireland. Vox Sang. 1999; 76(3): 175-180. doi: 10.1159/000031045
- Weiner OH, Alt M, Durr R, Noegel AA, Caselmann WH. Rapid and reproducible quantification of hepatitis C virus cDNA by fluorescence correlation spectroscopy. Digestion. 2000; 61(2): 84-89. doi: 10.1159/000007739
- Heller T, Hoofnagle J, Liang TJ, Rehermann B. Acute hepatitis C. Gastroenterology. 2009; 136(7): 2411. doi: 10.1053/j.gastro.2009.04.042
- Baumert TF, Wellnitz S, Aono S, et al. Antibodies against hepatitis C viruslike particles and viral clearance in acute and chronic hepatitis C. Hepatology. 2000; 32(3): 610-617. doi: 10.1053/jhep.2000.9876
- Tsatsralt-Od B, Takahashi M, Endo K, et al. Infection with hepatitis A, B, C, and delta viruses among patients with acute hepatitis in Mongolia. J Med Virol. 2006; 78(5): 542-550. doi: 10.1002/jmv.20574
- Nomura F, Itoga S. [Molecular diagnosis of hepatitis B virus and hepatitis C virus infection]. Rinsho Byori. 2002; 123: 31-36.
- Podzorski RP. Molecular testing in the diagnosis and management of hepatitis C virus infection. Arch Pathol Lab Med. 2002; 126(3): 285-290. doi: 10.1043/0003-9985(2002)126<0285:MTITDA>2.0.CO;2
- de Chassey B, Navratil V, Tafforeau L, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008; 4: 230. doi: 10.1038/msb.2008.66
- McCarthy MC, el-Tigani A, Khalid IO, Hyams KC. Hepatitis B and C in Juba, southern Sudan: results of a serosurvey. Trans R Soc Trop Med Hyg. 1994; 88(5): 534-536.
- El-Hazmi MM. Prevalence of HBV, HCV, HIV-1, 2 and HTLV-I/II infections among blood donors in a teaching hospital in the Central region of Saudi Arabia. Saudi Med J. 2004; 25(1): 26-33.
- Al Traif I, Al Balwi MA, Abdulkarim I, et al. HCV genotypes among 1013 Saudi nationals: a multicenter study. Ann Saudi Med. 2013; 33(1): 10-12. doi: 10.5144/0256-4947.2013.10
- Hamoudi W, Ali SA, Abdallat M, Estes CR, Razavi HA. HCV infection prevalence in a population recruited at health centers in Jordan. J Epidemiol Glob Health. 2013; 3(2): 67-71. doi: 10.1016/j.jegh.2013.02.003
- Suliman SM, Fessaha S, El Sadig M, et al. Prevalence of hepatitis C virus infection in hemodialysis patients in Sudan. Saudi J Kidney Dis Transpl. 1995; 6(2): 154-156.
- Sato S, Fujiyama S, Chikazawa H, Tanaka M. [Antibodies generated against hepatitis C virus (HCV) core antigen: IgM and IgA antibody against HCV core]. Nihon Rinsho. 1995; 53: 253-259.
- Taura Y, Fujiyama S, Kawano S, et al. Clinical evaluation of titration of hepatitis C virus core antibody and its subclasses. J Gastroenterol Hepatol. 1995; 10(3): 270-276.
- Pereira BJ, Milford EL, kirkman RL, et al. Prevalence of hepatitis C virus RNA in organ donors positive for hepatitis C antibody and in the recipients of their organs. N Engl J Med. 1992; 327(13): 910-915. doi: 10.1056/NEJM199209243271302
- Abacioglu YH, Bacaksiz F, Bahar IH, Simmonds P. Molecular evidence of nosocomial transmission of hepatitis C virus in a haemodialysis unit. Eur J Clin Microbiol Infect Dis. 2000; 19(3): 182-186.
- Abdel-Aziz F, Habib M, Mohamed MK, et al. Hepatitis C virus (HCV) infection in a community in the Nile Delta: population description and HCV prevalence. Hepatology. 2000; 32(1): 111-115. doi: 10.1053/jhep.2000.8438


