Research Article
Natural Plasmids in a Swiss Fermented Milk and Colostrum Product assessed by Microbiome Array
Silver Spring Sagl, Arzo-Mendrisio, Switzerland
*Corresponding author: Stefania Pacini, Silver Spring Sagl, Via Raimondo Rossi 24, Arzo-Mendrisio 6864, Switzerland, E-mail: info@bravo-europe.com
Received: October 1, 2019 Accepted: October 16, 2019 Published: October 24, 2019
Citation: Pacini S, Ruggiero M. Natural Plasmids in a Swiss Fermented Milk and Colostrum Product assessed by Microbiome Array. Madridge J Immunol. 2019; 3(2): 100-108. doi: 10.18689/mjim-1000123
Copyright: © 2019 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Natural plasmids are present in fermented foods ranging from yogurts to sourdoughs, sausages, kimchi or pickling crabs. In this study, we describe the natural plasmids present in a product obtained by fermentation of bovine milk and colostrum. We used a microarray consisting in a chip covered in short DNA sequences that are specific to target microorganisms for a total of approximately 12,000 species. As far as plasmids are concerned, the array yielded 48 iterations, each one corresponding to a unique plasmid target. We discuss the putative role of some of these plasmids in contributing to the known properties of probiotics in supporting health and longevity.
Keywords: Plasmids; Milk; Colostrum; Microbiome; Gene Transfer; Immunity.
Introduction
Foods and beverages fermented by microbes were among the first processed aliments our human ancestors consumed [1], and it is well accepted, since the observation by Metchnikoff, the “Father of Natural Immunity”, that microbes of fermented foods contribute to human health and longevity [2]. Todayʼs research on the role of the microbiota confirms the intuition of our ancestors who exploited the health benefits of fermented foods and beverages; we are learning that microbes are essential constituents of our bodies and, possibly, our minds since the brain itself has its own unique microbiota [3]. The role of the microbiota in determining health and longevity is now so embedded in popular culture that Vogue UK recently highlighted this concept with the following words “The human body contains many trillions of microbes. Their interactions create vitamins, break down our food, fight infections and communicate with our genes: without a microbiome we are royally buggered” [4]. This perception fits in the developing idea that healthy life expectancy (HALE) is different from life expectancy tout court; while global average life expectancy has increased in the past twenty years, HALE has not increased at the same pace. Decrease of mortality in children and adults is responsible for increased life expectancy, but it is a life affected by chronic diseases up to the point that respected Authors write of “morbidity expansion” [5]. It is increasingly accepted that a healthy microbiota is the key to healthy aging and life expectancy [6]. The “trillions of microbes” of the human microbiota harbor a proportional number of genes in their chromosomes as well as a huge number of plasmids carrying advantageous genetic information that can be transferred to other microbes and, possibly, human cells. Plasmids are made of double-strand DNA that is generally of small size and assumes a closed conformation often referred to as circular, even though the topology of DNA in plasmids may prove highly complex. Plasmids exist in nature in prokaryotes and in some eukaryotes; the genes present in plasmids provide cells with genetic advantages that favor survival and can be transferred to other cells. Since plasmids have been used for decades in laboratories for genetic engineering, it is important to distinguish between natural plasmids and their genetically-modified counterparts that are generated for research and clinical applications [7]. Natural plasmids are omnipresent in fermented ailments ranging from Bulgarian yogurts [8] to Thai sausages, Kimchi or pickling crabs [9], from Swedish and German sour doughs [10] to Indonesian dadih (fermented buffalo milk) [11]. We recently described the microbial composition of a Swiss fermented milk and colostrum product whose versions have been used in the nutritional approach to chronic conditions since 2014; in that study, we focused our attention on the composition in bacteriophages [12]. Our description was based on exhaustive genetic analysis using a novel type of microarray, the Axiom Microbiome Array, a tool for analysis that is “the most comprehensive microbial detection platform built to date” [13]. Using this extraordinarily potent array to characterize in fine detail the microbial composition of the fermented product quoted above, we noticed that its biodiversity was much higher than previously thought and comprised a significant number of plasmids. Here we describe the plasmids present in this fermented product and we discuss the putative role of some of these plasmids in conferring health supporting properties with particular reference to their role in HALE and detoxification.
Materials and Methods
The Axiom Microbiome Array was performed by Eurofins Microbiology Laboratories Inc. (New Berlin, Wisconsin, USA) on a commercially available product designated Freeze-Dried Bravo-Colostrum and Probiotic Complex (Silver Spring Sagl, Arzo-Mendrisio, Switzerland). The results presented in this paper refer to the same product whose composition in phages was recently described [12]. After fermentation of bovine milk and colostrum at room temperature for 48 hrs, the resulting fermented product undergoes the process of freeze drying. The characteristics of the product as far as nutrition facts and microbial composition are concerned are described in a recent paper [14]. The Axiom Microbiome array used for this analysis is a chip covered in short DNA sequences that are specific to certain target organisms and a total of approximately 12,000 species are included. For each target organism, the Microbiome Detection Analysis Software (MiDAS) looks at the ratio of probes that successfully bound to their target (Probes Observed) to the total number that could potentially bind (Probes Expected), and uses this to calculate the Initial Score, which is proportional to the likelihood of the target existing in the tested sample. Once the software identifies the target that it deems most likely to be present in the actual sample, the probes that correspond to the chosen target are removed and the process is repeated until adding new species no longer adds explanatory power to the softwareʼs model of the composition of the sample. The products of this iterative analysis are called Iterations. Probes are considered positive when presenting signal intensity higher than 99th percentile of the random control probe intensities with a percentage of more than 20% of target-specific probes detected. In other words, a target organism is considered present in the sample when the percentage of observed probes is greater than 20% of the expected probes. In the example provided by the Authors who first described this assay [13], DNA from vaccinia virus was detectable when there were 1,000 or more copies of the viral DNA in the examined sample. The number of probes observed at 1,000 copies was 78 out of 293 expected probes that is about 26%, and this value was deemed positive for the presence of the virus. Although this is not a quantitative assay, some sort of quantification can be deducted; in the example of the vaccinia virus DNA, the number of probes observed at 10,000 copies was higher than that observed at 1,000 copies as it was 148 out of 286 probes expected, or about 52%. The increase of one order of magnitude in the number of copies of vaccinia virus DNA was reflected by the increase from 26 to 52%. Such proportionality between that quantity of DNA and the percentage of observed copies is present in bacteria as well. When Shigella flexneri was tested, its DNA was positively detected at 100 genome copies and above, with a lower limit at 10 genome copies. The number of probes observed at 100 copies was 2,185 out of 7,398 probes expected, or about 29%, whereas the number of probes observed at 1,000 copies was 3,781 out of 8,009 probes expected, or about 47% [13]. The Axiom Microbiome Array is capable of accurately detecting members of mock microbial communities used for testing, achieving 100% accuracy in species identification; the specificity of the array yields precious information for diagnosis and surveillance of pathogens in food matrices, at the same time providing strain level detections since the array was designed to detect specific genomic regions of the sequences represented on the chip.
Results
The array highlighted the presence of a very high number of targets corresponding to microbial species that fit into the definition of probiotics and are members of the families of Streptococcaceae, Lactobacillaceae, Leuconostocaceae, Bifidobacteriaceae, and Thermaceae. Since the product contains a blend of lyophilized kefir grains and yogurt starter with live cultures that are used to ferment milk and colostrum, we hypothesize that fermentation by the complex microflora of the kefir grains contributed to the extreme biodiversity evidenced by the Axiom Microbiome Array. Consistent with this hypothesis, it was recently demonstrated that a typical fermented kefir contains hundreds of different microbial species; such a biodiversity is deemed responsible for the known health-supporting properties of kefirs that range from modulating the immune system to fighting carcinogenesis and allergies, from favoring wound healing to regulating cholesterol metabolism and blood pressure [15].
Table 1 shows the plasmid composition of the product. The array provided 48 iterations, each one corresponding to a unique target. The targets are ordered in descending sequence with those showing the highest percentage of observed probes at the top of the list. Although the assay is not quantitative, it can be deduced that higher percentages correspond to higher number of copies [13].
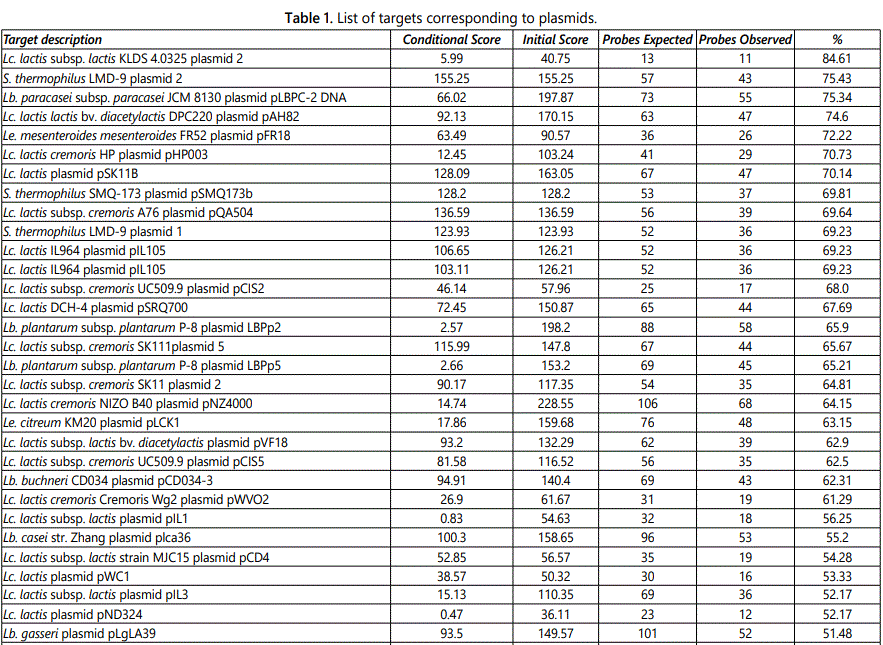
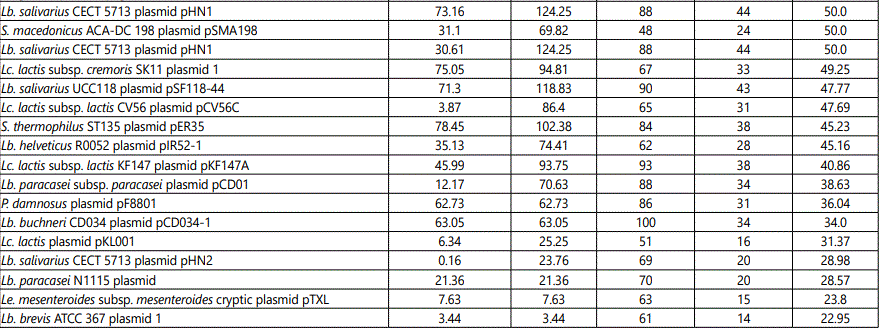
Table 2 shows the targets corresponding to Lactobacilli present in the product. The array identified the following Lactobacillus strains: paracasei, casei, helveticus, kefiranofaciens, plantarum, gallinarum, delbrueckii, rhamnosus, salivarium, acidophilus, farraginis and antri. The substrains are listed in the table.
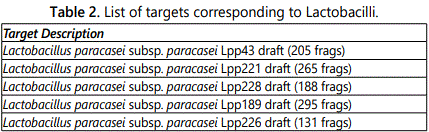
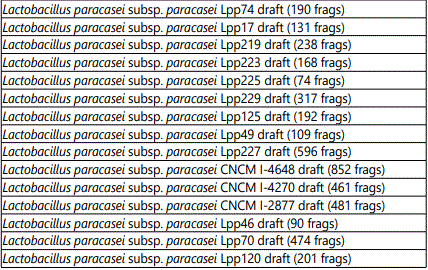
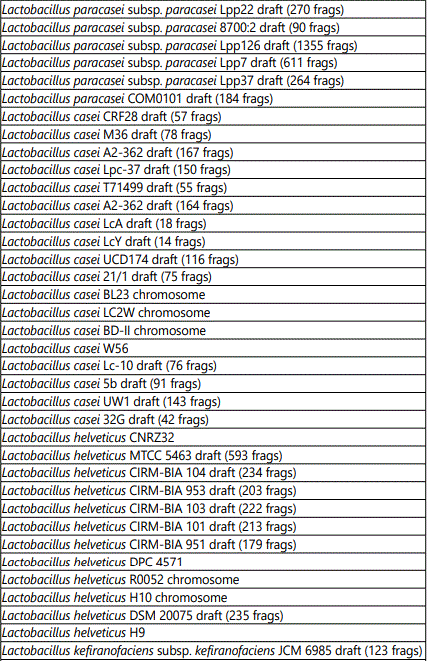
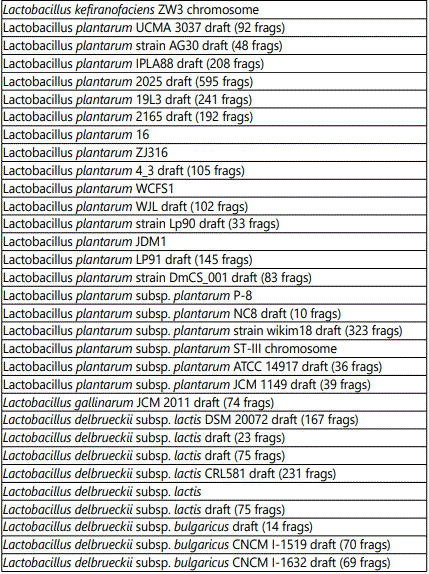
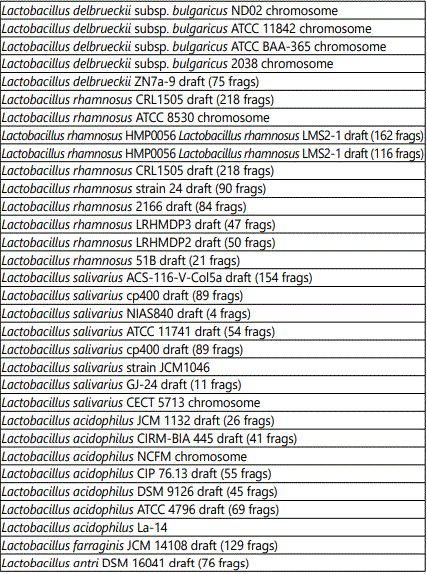
Table 3 shows the targets corresponding to Lactococcus lactis strains present in the product. The substrains identified by the array are listed in the table.

Discussion
Here, we discuss the main features of some of the plasmids evidenced by the Axiom Microbiome Array in the product object of this study as they potentially relate to human health, longevity and healthy life expectancy.
Lactobacillus (Lb.) casei strain Zhang plasmid plca 36
The probiotic Lb. casei Zhang was identified by screening of lactic acid bacteria isolated from samples of koumiss, a traditional home-made drink obtained from mareʼs milk by nomadic populations in China and Inner Mongolia [16,17]. Traditionally, consumption of koumiss is deemed beneficial for improving the symptoms of digestive diseases and of a wide range of chronic diseases, ranging from tuberculosis to bronchitis and anemia, thus suggesting that it may be effective in strengthening and modulating the immune system. Consistent with these anecdotic observation, it was demonstrated that Lb. casei Zhang has antipathogen, antioxidant, and immunomodulatory properties and is highly resistant to the acidic environment of the stomach as well as to the stress exerted by the components of bile [16,17]. Lb. casei Zhang has a 2,861,848-bp circular chromosome and a plasmid of 36-kb exhibiting a lower content of G+C in comparison with the chromosome [18]. Lb. casei Zhang has more phosphotransferase system (PTS)-related proteins than other Lactobacillus strains; significant redundancy of PTSs encoded in the chromosome of Lb. casei Zhang may be advantageous for the transport and use of a large panel of carbon sources, thus highlighting the flexibility and adaptability to different environments of this peculiar probiotic. Plasmid plca36 contains 44 predicted coding regions, and a precise function is assigned to 23 of them [18]. Of particular interest is a relBE toxin-antitoxin (TA) locus. TA loci or systems were originally described in low-copy-number plasmids; they have the function to maintain the plasmid stability by post-segregational elimination of plasmid-free daughter cells. However, bioinformatics and experimental evidence demonstrated that the TA modules are widely spread not only upon plasmids but also on bacterial chromosomes. The relBE TA module of plca36 may serve the role of allowing survival of the host under extreme nutritional stress as it works as a cell growth modulator which helps Lb. casei Zhang to deal with nutritional stress. Also a region encoding a cluster of conjugation genes (tra) is of interest since it shows high similarity and co-linearity with analogous regions of plasmids pWCFS103 and pMRC01 from Lb. plantarum and Lb. lactis, respectively. Comparative gene analysis revealed that plasmids from the genus Lactobacillus may have contributed to the environmental adaptation by providing carbohydrate and amino acid transporters. In summary, the genes encoded in plca36 appear to confer a significant advantage in surviving the harsh environmental conditions that are presumably encountered in Inner Mongolia. If presence of an advantageous plasmid from a Mongolian strain in a Swiss product were not strange enough, the Axiom Microbiome Array provided another element of microbiological curiosity; DNA from the chromosome of Lb. casei Zhang was not detected in the product. In other words, it appears that there is the plasmid but not the bacterium as if the plasmid travelled from Mongolia to Switzerland through innumerable passages in bacterial hosts of kefir grains sharing the genetic advantages originated in Mongolia with bacteria cultivated in Europe.
Lactobacillus (Lb.) buchneri CD034 plasmids pCD034-1 and pCD034-3
Lb. buchneri strains are known to improve stability of fermenting matrices upon oxygen exposure of conserved organic material thanks to their heterofermentative metabolism and the production of high levels of acetate [19]. Lb. buchneri CD034 possesses a circular chromosome and three additional plasmids two of which, pCD034-1 and pCD034-3, were detected by the Axiom Microbiome Array. In analogy with the case of Lb. casei Zhang, DNA from the chromosome of the bacterium was not observed; in this case, however, exotic transmigration of microbial DNA is probably not involved as Lb. buchneri is widely represented in Europe. The largest plasmid, pCD034-3, codes for 57 genes that comprise a putative polysaccharide synthesis gene cluster, whereas the functions of the two smaller plasmids, pCD034-1 and pCD034-2, are not fully elucidated [20]. In the examined product, the percentage of observed probes for pCD034-3 was 62% of expected probes, whereas the percentage was significantly lower for pCD034-1 (34%). Absence of the chromosomal DNA of the bacterium and of plasmid pCD034-2 seems to indicate that the fermented product retained only the advantageous genetic information contained in plasmids pCD034-1 and pCD034-3 that were probably selected by the peculiarities of kefir fermentation. Lb. buchneri is characterized by heterofermentative metabolism with high production of acetate and, in modern agriculture, these characteristics are exploited for grass silage as they result in improved stability upon oxygen exposure of the conserved plant material. The process of silage features three phases; at first, aerobic conditions are present during harvesting and preparation of green fodder. Then, proper ensiling begins after storage of the ensiled material under conditions that prevent aeration and lead to anaerobiosis. During the fermentation that follows, water soluble carbohydrates are used by lactic acid bacteria and lactic acid is produced with consequent acidification. When the silage is opened, aerobic conditions are restored as oxygen is available to the ensiled material. Since these aerobic conditions may then favor growth of spoiling microbes such as yeasts, fungi and other bacteria, Lb. buchneri is inoculated into the fresh plant biomass before ensiling so to preserve the stability of the material when aerobic conditions are restored at the opening of the silage [19]. Interestingly, similar conditions of aerobiosis followed by anaerobiosis and then by aerobiosis, occur during fermentation of the product described in this study, in particular considering the lower strata of the fermenting product that are not exposed to oxygen until the end of the process. Therefore, it can be assumed that acquisition of Lb. buchneri plasmids by other Lactobacilli occurred as means of exploiting an evolutionary advantage associated with the peculiar conditions of fermentation. In other words, the transferable genetic information of Lb. buchneri may contribute to the stability of the product object of this study just as they contribute to the stability of fermented plant biomasses in ensiling. What is peculiar is the fact that, in ensiling, Lb. buchneri is purposely added as inoculum, whereas in the product described in this study, its plasmids occur naturally as if natural evolution were well aware of what are now considered sophisticated methods of modern agriculture.
Lactococcus (Lc.) lactis, subsp. cremoris SK11 plasmids SK11B, 1, 2, 5
Strains of Lc. lactis carry plasmids that encode traits that are important for the dairy industry; a list of the targets corresponding to substrains of Lc. lactis evidenced by the Axiom Microbiome Array is reported in table 3. Among these, Lc. lactis subsp. cremoris SK11 is widely used in manufacture of fermented milk products. The Axiom Microbiome Array evidenced the presence of 4 plasmids of Lc. lactis subsp. cremoris SK11, namely SK11B, SK11 5, SK11 2, and SK11 1, here listed from the most to the least represented (Table 1). The significant over-representation of pSK11B is consistent with the hypothesis that this is a high-copy-number plasmid in comparison with the other three. It should be noticed, however, that other plasmids of different Lc. lactis subsp. cremoris strains were more represented than SK11B. Other plasmids pertaining to Lc. lactis subsp. cremoris strains identified by the array were; pHP003, pQA504, pCIS2, pNZ4000, pCIS5, and pWVO2 (Table 1). Many substrains of Lc. lactis subsp. cremoris have plasmids that encode traits responsible for catabolism of lactose, utilization of citrate, synthesis of proteases, production of bacteriocin, resistance to phages, synthesis of the exopolysaccharide, and, most important in this context, resistance to heavy metals [21]. Plasmid SK11B carries the gene aldC that codes for a protein of 221 amino acids with more than 90% identity to an alpha-acetolactate decarboxylase of the Lc. lactis IL-1403 chromosome. This enzyme leads to production of acetoin, a molecule that plays important roles in the regulation and detoxification of pyruvate metabolism through pyruvate dehydrogenase [21]. The enzyme is also involved in regulation of biosynthesis of branched-chain amino acids, a metabolic step of great importance for protein building. However, possibly more interesting, is the role that the enzyme may have in formation of a deep electron sink. The insertion sequence upstream of the gene aldC encoded by the plasmid could be responsible for over expression of the gene with the resulting creation of additional electron sinks. Creation of electron sinks may be instrumental in the processes of detoxification and it is postulated that electron sinks were at the basis of life evolution on Earth. According to Authors from the Centre National de la Recherche Scientifique of Marseille, France, a deep electron sink available to the last universal common ancestor before the divergence of Bacteria and Archaea was responsible for emergence of bioenergetic pathways that eventually led to aerobic respiration when oxygen became available [22].
Lactobacillus (Lb.) helveticus R0052 plasmid pIR52-1 Lb. helveticus was described for the first time in 1919 by Orla-Jensen and was named Thermo bacterium helveticum, where the prefix thermos referred to the high temperature utilized to produce the Emmental cheese from where the bacterium was originally isolated [23]. In addition to dairy products, Lb. helveticus is naturally present in the human intestine where it has a number of health-supporting properties. The bacterium successfully colonizes the gut because it has the ability to survive the acidic environment of the stomach and withstand the components of bile [24]. The ability to survive in an acidic environment explains how the bacterium can proliferate rapidly in fermenting milk because of its resistance to acid stress; rapid growth in milk is accompanied by expression of genes coding for proteolytic enzymes that lead to release of a great quantity of newly formed peptides, including bioactive peptides derived from cleavage of milk proteins, in particular casein. Lb. helveticus cell-wall proteolytic enzymes lead to generation of tripeptides, Ile-Pro-Pro (IPP) and Val-Pro-Pro (VPP), deriving from the hydrolysis of casein in milk [25]. These tripeptides have a number of health-supporting activities that may explain the role of the probiotic in promoting HALE. They inhibit the angiotensin converting enzyme and, therefore, exert a preventive/therapeutic effect on hypertension. They also improve vascular endothelial function, a key element in prevention of cardiovascular diseases and this effect is independent from their hemodynamic effect. A clinical study concluded that administration of a casein hydrolysate - that is analogous to the hydrolyzed casein naturally present in fermented milk containing Lb. helveticus - could help preventing cardiovascular diseases in hypertensive subject [26]. Another study [27], reported that both IPP and VPP stimulated beneficial adipogenic differentiation with effects similar to those induced by insulin with potential benefits in the presence of insulin resistance. Most important for increasing HALE and achieving optimal health, it was demonstrated that IPP and VPP inhibited cytokine-stimulated pro-inflammatory alterations such as reduction in adipokine levels and activation of the nuclear factor kappa B pathway [27]. These observation are of utmost importance if we consider that chronic inflammation is responsible for the discrepancy between the observed increase in life expectancy and the non-corresponding increase in HALE as discussed by Nunn et al. [28]. According to these Authors “although average global life expectancy has gone up, it does not seem to be due to an increase in HALE, and is certainly not approaching anything like that possible for a human. One key driver for morbidity expansion is lifestyle- induced inflammation” that leads to accelerate aging. The healthsupporting properties of Lb. helveticus encompass the ability to detoxify carcinogenic organic compounds and modulate the immune system [29]. As far as detoxification is concerned, one of the mechanisms consists in binding and eliminating carcinogenic compounds such as heterocyclic amines that derive from cooking fish or meat. Another mechanism consists in the formation of toroidal structures that bind carcinogenic and toxic metals such as cadmium or aluminum as it will be discussed later. As far as immune modulating properties are concerned, Lb helveticus strain R0052 works on several mechanisms associated with immune responses. Of particular interest are the effects on macrophages that may explain the efficacy of one version of the fermented product here described [30]. Milk fermented with Lb. helveticus or its cell-free supernatant stimulated macrophages in culture and modulated the proliferation of lymphocytes in vitro [29]. These observations demonstrate that the immune-modulating properties of Lb. helveticus are associated with generation of molecules that are present in the cell-free supernatant. Since 2011 we postulated that also peptides deriving from the activity of glycosidases may contribute to the immune-modulating properties of the product here described [30]. Lb. helveticus DSM 20075, well represented in the product object of this paper (Table 2), codes for beta-galactosidase [31], and it was demonstrated that the activity of this enzyme is a common first-stage modification of the three major subtypes of vitamin D binding protein to vitamin D binding protein-Macrophage Activating Factor (DBP-MAF) [32]. The immune-modulating properties of the Lb. helveticus strains present in the product here described lead to further intriguing propositions as far as their involvement in HALE is concerned. For example, Lb. helveticus strain R0052, efficiently inhibited the invasion of two different strains of C. jejuni in human colon cancer cells [33] at the same time protecting healthy cells from alterations of innate immunity [34]. In another study, oral administration of a probiotic containing Lb. helveticus reduced the symptoms of depression after myocardial infarction [35]. The same formulation reduced apoptosis in the limbic areas of the brain that are involved in the pathophysiology of depression, thus suggesting that strains of Lb. helveticus exert their influence on the gut-brain axis possibly normalizing the function of the central nervous system [36]. Lb. helveticus plasmids contribute to these health-supporting properties since they contain information associated with proteolytic activity and lactate production. Lb. helveticus R0052 has a plasmid with eight open reading frames (ORFs); four of these codes for proteins whose function is unknown. Non-coding elements control the number of copies of the plasmid and its maintenance [37].
Lactobacillus (Lb.) plantarum subsp. plantarum P-8 plasmid LBPp2
Lb. plantarum strains are able to survive the acidic environment of the human stomach and are resistant to bile, thus being able to successfully colonize the gut and exert probiotic functions such as balancing the microbiota and preventing hyperlipidemia [38]. The strain P-8 was isolated from a traditionally fermented dairy product in China and was described in a publication of 2012 [39]. The bacterium has a circular chromosome of 3.03 Mb and seven plasmids. Among these, LBPp2 is the most abundant and possesses eight genes involved in regulation of metabolism; five of these code for galactosidases, thus further reinforcing the concept of plasmids being responsible for DBP-MAF generation from milk vitamin D binding protein [30,32]. In addition, LBPp2 codes for transposase and topoisomerase. The latter enzyme is responsible, among other functions, for the formation of toroids of DNA that may have particular importance for detoxification. In plasmids where the topology of DNA is in the shape of toroids, metals with electropositive charge such as cadmium, mercury or aluminum bind to the electronegative backbone of DNA with formation of disc-shaped structures where the metal is surrounded by the DNA toroid [40]. This feature of plasmids may contribute to the overall detoxifying effect of one version of the product that we reported in 2017 [41].
Conclusions
Presence of genetic information outside chromosomes in bacterial cells was known well before the observation of Watson and Crick in 1953; researchers in the late 1940s observed how extra chromosomal genetic elements served to transmit traits that were advantageous for microbes to survive in hostile environments and a number of designations for these hereditary elements was proposed; choncriogenes, plasmagenes, pangenes and episomes were among the names used before Joshua Lederberg in 1952 coined the word plasmid to designate “any extra chromosomal hereditary element” [42]. Genetic and molecular characterization led to classify plasmids in five categories; fertility F-plasmids, resistance plasmids, virulence plasmids, degradative plasmids, and Col plasmids. Plasmids, just as microbes in general, were at first considered villains because they were identified as elements responsible for transmitting resistance to antibiotics. However, just as the perception of microbes has changed up to the point that fashion magazines praise their health-promoting properties, also the perception of plasmids is changing. These small, highly mobile, genetic elements make possible the horizontal transfer of genetic information that reacts and adapts to the ever-changing environment. This transferable information is essential for survival in the competitive natural environment and, if acquisition and transfer of antibiotic resistance is detrimental for humans infected by pathogenic bacteria, resistance to toxic carcinogenic metals may prove highly appreciated in todayʼs polluted environment [43]. The plasmids detected by the Axiom Microbiome Array in the Swiss fermented product here described are associated with a number of healthy properties that may be responsible for the effects observed in conjunction with the use of the product in the context of complementary nutritional approaches to chronic conditions [14,30,41,44,45]. According to the results presented here and in Pacini and Ruggiero [12], we feel that the product described in this study may be useful for improving hale as well as for synergizing with therapeutic strategies in chronic conditions associated with inflammation and exposure to toxicants. In addition, it is tempting to speculate that plasmids may have other functions in addition to those associated with their role as accessory information for probiotic microbes. For example, they could transfer genetic information to mammalian cells and this information may prove useful to withstand adverse environmental conditions such as those involving exposure to toxic metals. It is known since 1975 that a fraction of plasmid DNA may integrate in the nucleus of mammalian cells [46] and it was demonstrated that small plasmids can persist for hours in serum of healthy subjects [47]. Genetically-modified plasmids have been proposed as therapy for a number of diseases ranging from cancer [48] to non-treatable neurological disorders, such as ischemic stroke, Parkinsonʼs disease, Alzheimerʼs disease, and multiple sclerosis [49]. It is therefore conceivable that the vast array of natural plasmids detected in the product here described may contribute to its health-supporting properties that can be envisaged in the broader context of increasing HALE and promoting detoxification.
Disclosures
Marco Ruggiero is CEO of Silver Spring Sagl, the company producing the fermented product described in this paper. Stefania Pacini works as Quality Control responsible person for Silver Spring Sagl. Both had no prior knowledge of the results of the Axiom Microbiome Array that was independently performed by Eurofins Microbiology Laboratories Inc. (New Berlin Wisconsin, USA) on a commercially available lot as part of the companyʼs quality controls.
Advisory
No information in this paper is intended or implied to be a substitute for professional medical advice, diagnosis or treatment.
References
- Makwana M, Hati S. Fermented Beverages and Their Health Benefits. In: Grumezescu AM, and Holban AM (eds). Fermented Beverages. 2019; 5: 1-29. doi: 10.1016/B978-0-12-815271-3.00001-4
- Gordon S. Elie Metchnikoff: father of natural immunity. Eur J Immunol. 2008; 38(12): 3257-3264. doi: 10.1002/eji.200838855
- Branton WG, Ellestad KK, Maingat F, et al. Brain microbial populations in HIV/AIDS: α-proteobacteria predominate independent of host immune status. PLoS One. 2013; 8(1): e54673. doi: 10.1371/journal.pone.0054673
- Spicer K. What Is Klotho And Will It Really “Induce Age-Reversal”? Vogue UK. 2019: 203-222.
- Salomon JA, Wang H, Freeman MK, et al. Healthy life expectancy for 187 countries, 1990-2010: a systematic analysis for the Global Burden Disease Study 2010. Lancet. 2012; 380(9859): 2144-2162. doi: 10.1016/S0140-6736(12)61690-0
- Kim S, Jazwinski SM. The Gut Microbiota and Healthy Aging: A MiniReview. Gerontology. 2018; 64(6): 513-520. doi: 10.1159/000490615
- Shimamura M, Sato N, Morishita R. Experimental and clinical application of plasmid DNA in the field of central nervous diseases. Curr Gene Ther. 2011; 11(6): 491-500.
- Petrova P, Danova S, Stoyancheva G, et al. Isolation and Characterization of Plasmids from Strains of Streptococcus Thermophilus Bulgarian Yogurt Starters. Biotechnol Biotechnol Equip. 2014; 17(1): 105-113. doi: 10.1080/13102818.2003.10819203
- Auputinan P, Tragoolpua Y, Pruksakorn S, Thongwai N. Profiles of plasmids in lactobacilli isolated from fermented foods. Chiang Mai J Sci. 2011; 38(4): 648-652.
- Lönner C, Preve-Åkesson K, Ahrné S. Plasmid contents of lactic acid bacteria isolated from different types of sour doughs. Curr Microbiol. 1990; 20(4): 201-207. doi:: 10.1007/BF02089411
- Napitupulu R, Kanti A, Yunlineri T, Hardiningsih R. Plasmid DNA of Lactobacillus from Indonesian traditional fermented foods with interest in developing food grade host-vector system. Journal Microbiology Tropical (Indonesia). 2002; 1(2): 91-96.
- Pacini S, Ruggiero M. Phage composition of a fermented milk and colostrum product assessed by microbiome array; putative role of open reading frames. bioRxiv. 2019. doi: 10.1101/714154
- Thissen JB, Be NA, McLoughlin K, Gardner S, Rack PG, Shapero MH, Rowland RRR, Slezak T, Jaing CJ. Axiom Microbiome Array, the next generation microarray for high-throughput pathogen and microbiome analysis. PLoS One. 2019; 14(2): e0212045. doi: 10.1371/journal.pone.0212045
- Antonucci N, Pacini S, Ruggiero M. Use of an Extremely Biodiverse Probiotic and a Supplement Based on Microbial Chondroitin Sulfate is Associated with a Significant Decrease of Serum Free Kappa Light Chains as well as a Trend Toward Normalization of Kappa/Lambda Ratio and of Plasma Cell Bone Marrow Infiltration in a Case of Multiple Myeloma. Am J Immunol. 2019; 15: 5-9. doi: 10.3844/ajisp.2019.5.9
- Bourrie BC, Willing BP, Cotter PD. The Microbiota and Health Promoting Characteristics of the Fermented Beverage Kefir. Front Microbiol. 2016; 7: 647. doi: 10.3389/fmicb.2016.00647
- Wu R, Wang L, Wang J, et al. Isolation and preliminary probiotic selection of lactobacilli from koumiss in Inner Mongolia. J Basic Microbiol. 2009; 49(3): 318-326. doi: 10.1002/jobm.200800047
- Zhang W, Yu D, Sun Z, et al. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J Bacteriol. 2010; 192(19): 5268-5269. doi:10.1128/JB.00802-10
- Zhang W, Yu D, Sun Z, et al. Complete nucleotide sequence of plasmid plca36 isolated from Lactobacillus casei Zhang. Plasmid. 2008; 60(2): 131-135. doi: 10.1016/j.plasmid.2008.06.003
- Eikmeyer FG, Heinl S, Marx H, Pühler A, Grabherr R, Schlüter A. Identification of Oxygen-Responsive Transcripts in the Silage Inoculant Lactobacillus buchneri CD034 by RNA Sequencing. PLoS One. 2015; 10(7): e0134149. doi:10.1371/journal.pone.0134149
- Heinl S, Wibberg D, Eikmeyer F, et al. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol. 2012; 161(2): 153-166: doi: 10.1016/j.jbiotec.2012.03.007
- Siezen RJ, Renckens B, van Swam I, et al. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl Environ Microbiol. 2005; 71(12): 8371-8382. doi: 10.1128/AEM.71.12.8371-8382.2005
- Ducluzeau AL, van Lis R, Duval S, et al. Was nitric oxide the first deep electron sink? Trends Biochem Sci. 2009; 34(1): 9-15. doi: 10.1016/j.tibs.2008.10.005
- Sabri M, Naser KE, Hagen MV, et al. Lactobacillus suntoryeus Cachat and Priest 2005 is a later synonym of Lactobacillus helveticus (Orla-Jensen 1919) Bergey et al. 1925 (Approved Lists 1980). Int J Syst Evol Microbiol. 2006; 56: 355-360. doi: 10.1099/ijs.0.64001-0
- Fontana A, Falasconi I, Molinari P, et al. Genomic Comparison of Lactobacillus helveticus Strains Highlights Probiotic Potential. Front Microbiol. 2019; 10: 1380. doi: 10.3389/fmicb.2019.01380
- Cremonesi P, Chessa S, Castiglioni B. Genome sequence and analysis of Lactobacillus helveticus. Front Microbiol. 2013; 3: 435. doi: 10.3389/fmicb.2012.00435
- Hirota T, Ohki K, Kawagishi R, et al. Casein Hydrolysate containing the Antihypertensive Tripeptides Val-Pro-Pro and Ile-Pro-Pro Improves Vascular Endothelial Function Independent of Blood Pressure–Lowering Effects: Contribution of the Inhibitory Action of Angiotensin-Converting Enzyme. Hypertens Res. 2007; 30(6): 489-496. doi: 10.1291/hypres.30.489
- Chakrabarti S, Wu J. Milk-derived tripeptides IPP (Ile-Pro-Pro) and VPP (Val-Pro-Pro) promote adipocyte differentiation and inhibit inflammation in 3T3-F442A cells. PLoS One. 2015; 10(2): e0117492. doi: 10.1371/journal.pone.0117492
- Nunn AV, Guy GW, Bell JD. The quantum mitochondrion and optimal health. Biochem Soc Trans. 2016; 44(4): 1101-1110. doi: 10.1042/BST20160096
- Taverniti V, Guglielmetti S. Health-Promoting Properties of Lactobacillus helveticus. Front Microbiol. 2012; 3: 392. doi: 10.3389/fmicb.2012.00392
- Pacini S, Punzi T, Morucci G, Ruggiero M. Macrophages of the mucosaassociated lymphoid tissue (MALT) as key elements of the immune response to vitamin d binding protein-macrophage activating factor. Italian Journal of Anatomy and Embryology. 2011; 116(1): 136. doi: 10.13128/IJAE-10160
- Kittibunchakul S, Pham ML, Tran AM, Nguyen TH. β-Galactosidase from Lactobacillus helveticus DSM 20075: Biochemical Characterization and Recombinant Expression for Applications in Dairy Industry. Int J Mol Sci. 2019; 20(4): 947. doi: 10.3390/ijms20040947
- Uto Y, Yamamoto S, Mukai H, et al. β-Galactosidase treatment is a common first-stage modification of the three major subtypes of Gc protein to GcMAF. Anticancer Res. 2012; 32(6): 2359-2364.
- Wine E, Gareau MG, Johnson-Henry K, Sherman PM. Strain-specific probiotic (Lactobacillus helveticus) inhibition of Campylobacter jejuni invasion of human intestinal epithelial cells. FEMS Microbiol Lett. 2009; 300(1): 146-152. doi: 10.1111/j.1574-6968.2009.01781.x
- Jandu N, Zeng ZJ, Johnson-Henry KC, Sherman PM. Probiotics prevent enterohaemorrhagic Escherichia coli O157: H7-mediated inhibition of interferon-gamma-induced tyrosine phosphorylation of STAT-1. Microbiology. 2009; 155: 531-540. doi: 10.1099/mic.0.021931-0
- Arseneault-Bréard J, Rondeau I, Gilbert K, et al. Combination of Lactobacillus helveticus R0052 and Bifidobacterium longum R0175 reduces post-myocardial infarction depression symptoms and restores intestinal permeability in a rat model. Br J Nutr. 2012; 107(12): 1793-9. doi: 10.1017/S0007114511005137
- Girard SA, Bah TM, Kaloustian S, et al. Lactobacillus helveticus and Bifidobacterium longum taken in combination reduce the apoptosis propensity in the limbic system after myocardial infarction in a rat model. Br J Nutr. 2009; 102(10): 1420-1425. doi: 10.1017/S0007114509990766
- Hagen KE, Tramp CA, Altermann E, Welker DL, Tompkins TA. Sequence analysis of plasmid pIR52-1 from Lactobacillus helveticus R0052 and investigation of its origin of replication. Plasmid. 2010; 63(2): 108-117. doi: 10.1016/j.plasmid.2009.12.004
- Song Y, He Q, Zhang J, et al. Genomic Variations in Probiotic Lactobacillus plantarum P-8 in the Human and Rat Gut. Front Microbiol. 2018; 9: 893. doi: 10.3389/fmicb.2018.00893
- Bao Y, Zhang Y, Li H, et al. In vitro screen of Lactobacillus plantarum as probiotic bacteria and their fermented characteristics in soymilk. Ann Microbiol. 2012; 62: 1311-1320. doi: 10.1007/s13213-011-0377-4
- Samson J, Varotto A, Nahirney PC, Toschi A, Piscopo I, Drain CM. Fabrication of metal nanoparticles using toroidal plasmid DNA as a sacrificial mold. ACS Nano. 2009; 3(2): 339-344. doi: 10.1021/nn800758n
- Blythe J, Ruggiero M, Pacini S. Intermittent fasting and probiotic yogurt consumption are associated with reduction of serum alpha-Nacetylgalactosaminidase and increased urinary excretion of lipohilic toxicants. Madridge J Immunol. 2017; 1(1): 23-27. doi: 10.18689/mjim1000107
- Lederberg J. Plasmid (1952-1997). Plasmid. 1998; 39(1): 1-9. doi: 10.1006/plas.1997.1320
- Chellaiah ER. Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: a minireview. Appl Water Sci. 2018; 8: 154. doi: 10.1007/s13201-018-0796-5
- Ruggiero M, Ward E, Smith R, et al. Oleic Acid, deglycosylated vitamin D-binding protein, nitric oxide: a molecular triad made lethal to cancer. Anticancer Res. 2014; 34(7): 3569-3578.
- Branca JJ, Pacini S, Ruggiero M. Effects of Pre-surgical Vitamin D Supplementation and Ketogenic Diet in a Patient with Recurrent Breast Cancer. Anticancer Res. 2015; 35(10): 5525-5532.
- Goebel W, Schiess W. The fate of a bacterial plasmid in mammalian cells. Mol Gen Genet. 1975; 138(3): 213-23. doi: 10.1007/bf00269348
- Rozenberg-Arska M, Salters EC, van Strijp JA, Hoekstra WP, Verhoef J. Degradation of Escherichia coli chromosomal and plasmid DNA in serum. J Gen Microbiol. 1984; 130(1): 217-222. doi: 10.1099/00221287-130-1-217
- Teo PY, Cheng W, Hedrick JL, Yang YY. Co-delivery of drugs and plasmid DNA for cancer therapy. Adv Drug Deliv Rev. 2016; 98: 41-63. doi: 10.1016/j.addr.2015.10.014
- Shimamura M, Sato N, Morishita R. Experimental and clinical application of plasmid DNA in the field of central nervous diseases. Curr Gene Ther. 2011; 11(6): 491-500.


