Mini-Review Article
The Role of LncRNAs in Immunity
1Investigator, Department of Microbiology and Immunology, School of Medicine, Stanford University, CA, USA
2Investigator, Carnegie Institute of Washington, Stanford, CA, USA
*Corresponding author: Yan Wang, Investigator, Department of Microbiology and Immunology, School of Medicine, Stanford University, CA, USA, Tel: 6507208364, E-mail: ywang15@stanford.com
Received: February 24, 2016 Accepted: April 2, 2016 Published: April 5, 2016
Citation: Wang Y, Jin H. The Role of LncRNAs in Immunity. Madridge J Immunol. 2016; 1(1): 1-4. doi: 10.18689/mjim-1000101
Copyright: © 2016 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Studies of genome suggest that the protein coding regions represent only around 1-2% of the genome. Recent studies indicate that the majority of the genome is transcribed into non-coding RNAs (ncRNAs), which play a key role in gene expression regulation. They are roughly classified into either short ncRNAs with less than 200 nucleotides or long ncRNAs (lncRNAs) with more than 200 nucleotides. While the short ncRNAs, have been well studied, lncRNAs have only started to shed their lights recently. Except having regulation functions for various diseases, lncRNAs have been shown to play a role in immune-response and immune cells. In this mini-review, we summarized the subtypes of lncRNAs, and further list the mechanism of how lncRNAs play their functions in immune systems.
Keywords: Immunity; Genome; Rib nucleoprotein; Phosphorylation.
Abbreviations: ncRNAs: Non-coding RNAs; lncRNA: Long Non-coding RNAs; miRNA: MicroRNA; siRNA: Small Interfering RNA; piRNA: Piwi-interacting RNA; SLE: Systemic Lupus Erythematosus; lincRNAs: Intergenic long Noncoding RNAs; AID: Autoimmune Disease; satRNAs: Satellite RNAs
Introduction
LncRNAs have been extensively expressed in cells. By deep sequencing analyzing, they are shown to be cell type and tissue type specific at different developmental stages. For instance, some lncRNAs are shown specifically expressed in pancreatic cancer, and therefore could be considered as markers [1]. Researches showed that knock down or block the lncRNA could regulate its targeted gene expression. Theoretically, lncRNAs contain domains that allow them to bind to proteins or RNA/DNA sequences, thus they could control gene expression. The main way is regulating expression of neighboring genes, or by interacting with chromatin at remote locations across multiple chromosomes to regulate the expression of multiple genes.
Human body precisely controls various cell types involved in the immune systems. When challenged by antigens, the innate immune cells will very quickly response; then usually after three days; the adaptive immune response will start. How to control and regulate different immune cells needs a system of regulation. Recent studies showed that immune cells could also be regulated by lncRNAs. In addition, immune related diseases are controlled by lncRNAs in both human and plants. The most well studied case is autoimmune disease (AID). In this mini review, the above topics will be discussed.
What's the lncRNA
RNA is traditionally considered as an intermediate bridge during protein translation process. However, the amount of these coding RNA only constitutes 1.5% of total RNA [2]. The percentage of non-coding RNA counts for more than 98% of total transcripts [3]. Increasing data showed that noncoding portions of RNA have been broadly involved in regulating the complexity of organisms. And the complexity of RNA sequence increases the ability of cells to store the huge information in genome [4,5]. This large group of ncRNA includes microRNA (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA) and long non-coding RNAs (lncRNA).
Unlike small noncoding RNAs, which have widely studied, the importance of lncRNAs has only been recognized recently [6]. However, it has started to show its potential: in human, lncRNAs are considered to be located in more than 50,000 different transcripts [7] of which, the number is nearly doubled than coding RNAs. In plant, lncRNAs are also shown largely involved in gene expression; around 6480 lncRNAs are related in gene profile of Arbidopsis thaliana [8].
Subtypes of lncRNAs
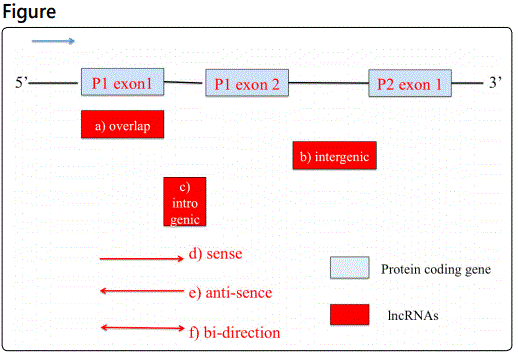
As one of the regulators for gene expression, lncRNAs could be grouped by its position of their target genes: (a) overlap, when it share same sequence with protein coding transcripts; (b) intergenic, when the lncRNA located between the coding regions of two protein; (c) intronic, when it sits in between two genes of one protein [9]. In addition to the position, the directions of transcripts and lncRNA also play an essential function for lncRNA's regulation. Therefore, it could be further divided to (d) sense; (e) antisense and (f) bi-direction three types (Figure 1) [10].
Due to these complexities of the lncRNAs' position as well as their transcripts direction, the functions of lncRNAs and their mechanisms are very different, and it has been shown hugely involved in various pathways and in many types of diseases.
Functions of lncRNAs
Although the study of lncRNA started from recent years, it has been shown the important functions in different types of diseases. In heart failure, lncRNAMhrt could inhibit chromatin targeting and Brg1 regulated gene expression for chromatin remodeling [11]. LncRNAs have also been shown a role in cancer; for instance, in pancreatic cancer, tissue specific lncRNAs have been identified [1]. Specifically, such as lncRNAsH19, HOTAIR, are unregulated in tumorigenic pancreatic tissue and further cause cell proliferation, migration and invasion [12,13]. Besides, in lymph node-negative breast cancer, the profile of lncRNAs could be use as prognostic markers [14]. In brain, if stroke happens, the expression profile of lncRNAs also changed [15], the same group also showed that lncRNAFosDT could further promote the ischemic brain injury by interacting with chromatin-modifying proteins [16].
Although the functions of lncRNAs in immune response have not been studied extensively as in cancer, it has been shown that immune diseases are also related to lncRNAs. Specifically, autoimmune disease (AID) is the one type of diseases, which has been relatively deeply studied. In 2014, Hrdlickova et al. showed compared AID to the whole genome, there was a significant difference in lncRNAs genome mapping loci of immune cells [17]. In the same year, Shi et al. showed that in Systemic Lupus Erythematosus (SLE), a prototypic system autoimmune disease, a lncRNA located on chromosome 6q25.3 has been down-regulation in patient, comparing to the gene expression of healthy control, which indicate the lncRNA could has a distinct function [18,19].
LncRNA is not only related to specific types of disease, it also regulates different immune cell types. In natural killer cells (NK cells), the over expression of myeloid zinc finger 1 (MZF-1) could decrease KIR expression, which is closely related to development of NKs. One lncRNA specifically expressed in NK cells, and was antisense to KIR has been found in progenitor cells or pluripotent cells could regulate KIR expression at the molecular level and further affected the development and differentiation of NK cells [20]. This lncRNA related regulation could balance the number of NKs in immune responses. In dendritic cell (DC), the lnc-DC was extensively expressed. The capacity of DC to present antigen to T cells and itself differentiation was reduced when the lncRNA was knockdown; this was because lnc-DC could promote the post-translational modification of STAT3 [21]. In addition to the examples above, intergenic long noncoding RNAs (lincRNAs), showed that during T cell differentiation, the cluster of lincRNA changed dynastically. Specifically, lincR-Ccr2-5'AS played a role for migration of Th2 cells [22]. The expression level of lincR-Ccr2-5'AS was controlled by GATA-3, this was a transcription factor and essentially for the differentiation of Th2 cells. Therefore, without GATA-3, the level of lincR-Ccr2-5'AS would be down regulated and blocked the development of Th2 cells. For macrophages, lncRNAPtprj-as1 was shown highly enriched, and had antisense direction to Ptprj [23]. Using Clustal W alignment, RVista and TRANSFAC, the transcription factor-binding sites were identified [23], which further showed the specificity of this lncRNA.
Not only in animals, in plant, lncRNAs also showed critical functions in plant immunity. One good example is satellite RNAs (satRNAs), which are 300-400bp long and therefore be considered as lncRNA, was shown a role between virus and host plant [24]. Specifically, this satRNA could improve the defense response to viruses by blocking the virus infection from plant's defense system [25,26]. In cucumber mosaic virus induced infection, satRNA- derived siRNA can specifically bind to the 3'UTR of virus RNA and further degrade it, therefore protect the plant from infection [25].
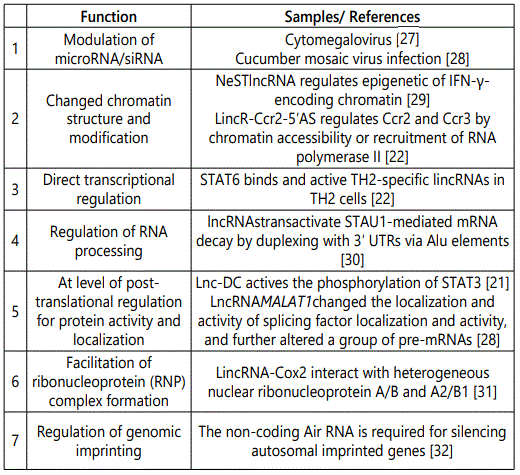
As we indicated above, lncRNAs have various locations and directions; therefore, it is reasonable that lncRNA could regulate the target gene by different mechanisms. Here, we summary the different mechanisms of how lncRNAs regulate their target genes (Table 1). Most of samples are listed using cases related to immune response.
Conclusion
As a regulator of gene expression, lncRNAs have started to show their important functions recently. Considering non-coding RNAs are accounts for the majority (above 95%) of genome, the function and mechanism of lncRNAs we know at the moment still very few. Both animals and plants require very precise control for their life; therefore, the response to the environment such as our immune response needs to be tightly controlled. As there are extensively genome information be stored in the cell, the regulation and functions of non-coding RNAs, such as lncRNA should be paid more attentions.
Conflicts of interest
Authors don't have any conflict of interest.
Acknowledgments
We thank Dr. Heng Zhao (Stanford University) and Dr. Pei Han for their suggestions and discussion when we did the lncRNA work.
References
- Peng JF, Zhuang YY, Huang FT, Zhang SN. Noncoding RNAs and pancreatic cancer. World J Gastroenterol. 2016; 22: 801-814. doi: 10.3748/wjg.v22.i2.801
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005; 309: 1559-1563. doi: 10.1126/science.1112014
- Ewan Birney, John A. Stamatoyannopoulos, Anindya Dutta, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447: 799-816. doi: 10.1038/nature05874
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004; 5: 316-323. doi: 10.1038/nrg1321
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011; 43: 904-914. doi: 10.1016/j.molcel.2011.08.018
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013; 12: 847-865. doi: 10.1038/nrd4140
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015; 47: 199-208. doi: 10.1038/ng.3192
- Liu J, Jung C, Xu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012; 24: 4333-4345. doi: 10.1105/tpc.112.102855
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015; 22: 5-7. doi: 10.1038/nsmb.2942
- Anamaria Necsulea, Magali Soumillon, Maria Warnefors, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014; 505: 635-640. doi: 10.1038/nature12943
- Pei Han, Wei Li, Chiou-Hong Lin, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014; 514: 102-106. doi: 10.1038/nature13596
- Matouk IJ, Raveh E, Abu-lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014; 1843: 1414-1426. doi: 10.1016/j.bbamcr.2014.03.023
- Kristina P Sørensen, Mads Thomassen, Qihua Tan, et al. Long non-coding RNA expression profiles predict metastasis in lymph node-negative breast cancer independently of traditional prognostic markers. Breast Cancer Res. 2015; 17: 55. doi: 10.1186/s13058-015-0557-4
- Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012; 43: 2800-2802. doi: 10.1161/STROKEAHA.112.669465
- Mehta SL, Kim T, Vemuganti R. Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated ChromatinModifying Proteins. J Neurosci. 2015; 35: 16443-16449. doi: 10.1523/JNEUROSCI.2943-15.2015
- Barbara Hrdlickova, Vinod Kumar, Kartiek Kanduri, et al. Expression profiles of long non-coding RNAs located in autoimmune diseaseassociated regions reveal immune cell-type specificity. Genome Med. 2014; 6: 88. doi: 10.1186/s13073-014-0088-0
- Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J Immunol Res. 2015; 2015: 848790. doi: 10.1155/2015/848790
- Lihua Shi, Zhe Zhang, Angela M. Y, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One. 2014; 9: e93846. doi: 10.1371/journal.pone.0093846
- Wright PW, Huehn A, Cichocki F, et al. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun. 2013; 14: 427-433. doi: 10.1038/gene.2013.36
- Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014; 344: 310-313. doi: 10.1126/science.1251456
- Gangqing Hu, Qingsong Tang, Suveena Sharma, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013; 14: 1190-1198. doi: 10.1038/ni.2712
- Richa K Dave, Marcel E Dinger, Megan Andrew, et al. Regulated expression of PTPRJ/CD148 and an antisense long noncoding RNA in macrophages by proinflammatory stimuli. PLoS One. 2013; 8: e68306. doi: 10.1371/journal.pone.0068306
- Shimura H, Masuta C. Plant subviral RNAs as a long noncoding RNA (lncRNA): analogy with animal lncRNAs in host-virus interactions. Virus Res. 2016; 212: 25-29. doi: 10.1016/j.virusres.2015.06.016
- Hui Zhu, Cheng-Guo Duan, Wei-Na Hou, et al. Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3' untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. J Virol. 2011; 85: 13384-13397. doi: 10.1128/JVI.05806-11
- Kiran Zahid, Jian-Hua Zhao, Neil A. Smith, et al. Nicotiana small RNA sequences support a host genome origin of cucumber mosaic virus satellite RNA. PLoS Genet. 2015; 11: e1004906. doi: 10.1371/journal.pgen.1004906
- Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014; 15: 484-491. doi: 10.1038/ni.2887
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010; 39: 925-938. doi: 10.1016/j.molcel.2010.08.011
- Gomez JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γlocus. Cell. 2013; 152: 743-754. doi: 10.1016/j.cell.2013.01.015
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011; 470: 284-288. doi: 10.1038/nature09701
- Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013; 341: 789-792. doi: 10.1126/science.1240925
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002; 415: 810-813. doi: 10.1038/415810a


