Research Article
Construction of Epitope-based Vaccine for Avian Reovirus
Running title: Epitopes-based vaccine for avian reovirus
1Department of Biological Sciences, Alabama State University, Montgomery, AL, 36101, USA
2Associate Vice President for Academic Operations, California State University, Fullerton, CA, 92831, USA
3Department of Poultry Science, Auburn University, AL 36849, USA
*Corresponding author: Hongzhuan Wu, Professor, Department of Biological Sciences, Alabama State University, Montgomery, AL, 36101, USA, E-mail: hwu@alasu.edu
Received: July 20, 2018 Accepted: September 5, 2018 Published: September 11, 2018
Citation: Wu H, Smith L, Scissum-Gunn K,Giambrone JJ, Robertson BK, Villafane R. Construction of Epitope-based Vaccine for Avian Reovirus. Madridge J Clin Res. 2018; 2(1): 50-54. doi: 10.18689/mjcr-1000109
Copyright: © 2018 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Epitope-based vaccine prototypes can stimulate protective immune responses against viruses. Identification of protective epitopes by traditional methods such as phage display is time and labor intensive. Such methods rely upon activity of epitope regions through empirical means for identification. Bioinformatics enables researchers to apply predictive approaches to identify protective epitopes. Software programs can predict epitopes by computer-driven pattern matching algorithms or analysis of conserved protein sequences for homology with known epitopes, however, such database services are expensive and limited to mouse and human pathogens. Avian reovirus (ARV) σ C protein is the main immunogenic surface protein of ARV. In this study, we developed a protocol for epitope prediction for an avian virus. Our predictions are based upon: (1) 2D and 3D structural analysis, (2) PROSITE glycosylation patterns, (3) protein sequence homology and alignments, and (4) hydrophobic index. Using this approach, we predicted 3 regions within the σ C protein that harbor putative protective epitopes. Then, we expressed 10 different size of recombinant σ C protein in a yeast expression system; the subject cell-neutralization of these recombinant proteins with avian reovirus S1133 strain in chicken fibroblast cells (CEF) not exactly collaborate our prediction.
Keywords: Reovirus; Sigma C; Epitopes; Bioinformatics.
Introduction
Avianreovirus (ARV) and mammalian reovirus (MRV) belong to the genus Orthoreovirus. Both share physical-chemical and morphological characteristic, including segmented genomes consisting of 10 genome segments of double-stranded (ds) RNA. The RNA is packaged into a non-enveloped icosahedral double capsid [1]. The genomic segments can be separated by polyacrylamide gel electrophoresis (PAGE) into three size classes, L (large), M (medium), and S (small). However, ARV differs from its mammalian counterpart in its lack of haemagglutination activity [2], ability to induce cell fusion [3, 4], and association with naturally occurring pathological conditions [5]. ARV is an important cause of diseases in poultry. In particular, reovirus-induced arthritis, chronic respiratory diseases, and malabsorption syndrome [6-9], provoke considerable economic losses.

All ARV-encoded proteins, including 10 structural and 4 nonstructural proteins, have been demonstrated [10, 11]. Protein sigma C, encoded by the S1 gene [12], is the target for type-specific neutralizing antibodies [13]. Recently, ARV sigma A encoded by the S2 gene [14] has been identified as a double-stranded RNA (dsRNA) binding protein [14, 15] and possible involvement in resistance to interferon [16]. Another protein of ARV, Sigma NS, encoded by the S4 gene [17], has been reported for its single-stranded RNA (ssRNA) binding activity [18]. To date, no investigations regarding the epitope mapping of anti-sigma C monoclonal antibodies have been reported.
Among mammalian reoviruses (MRVs), there are three structural proteins shown to carry neutralizing epitopes: σ1, σ2, and λ2. Hayers et al showed that σ1 protein carried type-specific neutralizing epitopes, whereas σ2 and λ2 carried group-specific neutralizing epitopes. ARV-encoded proteins, including at least 10 structural proteins (λA, λB, λC, µA, µB, µBC, µBN, σC, σA, and σB), and 4 nonstructural proteins (µNS, P10, P17, and σNS) have been demonstrated. Protein σC, encoded by the S1 gene, shows noticeably higher divergence than other σ-class proteins and is a cell attachment protein and apoptosis inducer. Protein σC is also the target for type-specific neutralizing antibodies while antibodies against σB are group-specific. ARV σA encoded by the S2 gene has been identified as a double-stranded RNA (dsRNA) binding protein and possible involvement in resistance to interferon. Among avian reoviruses, structure-based sequence alignment of the ARV sigma C and mammalian reovirus (MRV) type 3sigma 1 have indicated the presence of heptad repeats and a triple alpha-helical coiled-coil structure in N-terminal region. In addition, crystallographic studies found that the carboxy terminal globular domain of ARV sigma Chas a similar overall topology to that of MRV type 3 sigma 1. It has been shown that MRV sigma 1 protein recognizes the receptor of M cells (α-2-3 linked sialic acid) that facilitates penetration of antigens into intestinal Peyerʼs patches. However, there is little information on the ARV σC epitopes involved in the stimulation of host protective immunity. Moreover, current Bio-software are mostly mammalian-based and not commercial available, limited further bioinformatic analysis to its epitope region. In this study we first reported prediction of potential epitope regions on the sigma C protein of avian reovirus strain S1133 by bioinformatic analysis, S.pombe- expressed deletion fragments of sigma C was produced, and its neutralizing ability with polyclonal antibody against avian reovirus in chicken embryo cells was tested.
Materials and Methods
Bioinformaticsʼ analysis of sigma C protein
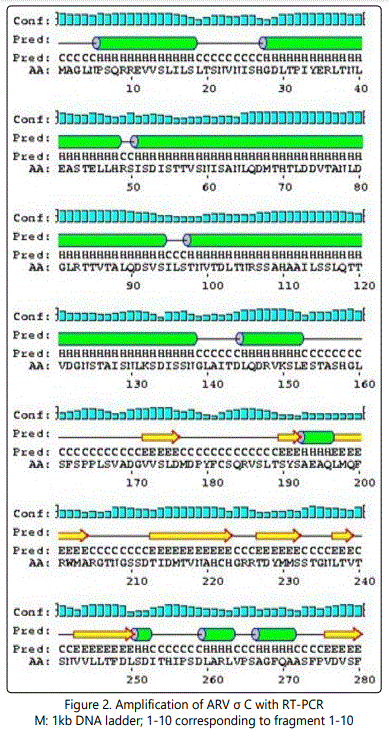
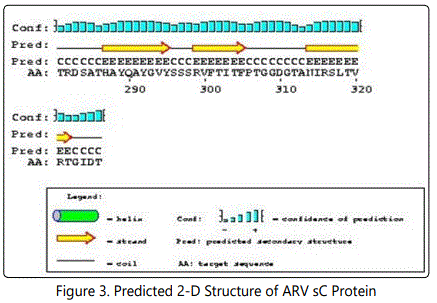
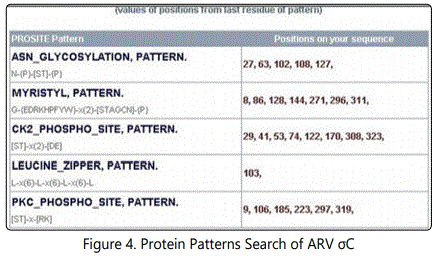
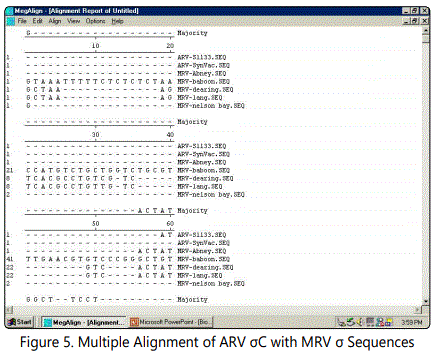
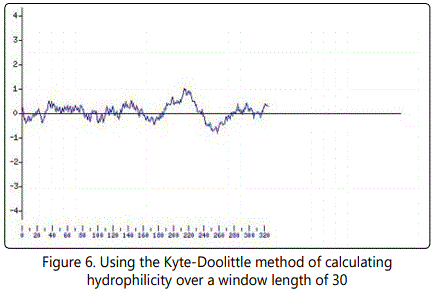
The following structure were compared:(1) 2D and 3D structural analysis, (2) PROSITE glycosylation patterns, (3) protein sequence homology and alignments, and (4) hydrophobic index. Using this approach, we predicted 3 regions within the σ C protein that harbor putative protective epitopes. Briefly, nucleotide and deduced amino acid sequences as well as the possible secondary structure of sigma C proteins were aligned and analyzed with a DNASTAR software package (DNASTAR Inc., Madison, WI, USA).The nucleotide sequence (981bp) of the full-length sigma C-encoding gene among ARV isolates were aligned, corresponding to residues 1-327 of the sigma C protein. The cDNA sequences and predicted amino acid sequences of the sigma C were used in paired identity analysis to determine the extent of nucleotide and amino acid sequence identity and divergence. The alignments were further analyzed with phylogeny program.
Gene expression and protein purification
The sigma C open reading frame was amplified by reverse-transcriptase polymerase chain reaction (RT-PCR). Primers for amplification of the 10 putative epitope region of σ C were obtained from Invitrogen, Inc. Carlsbad, CA.
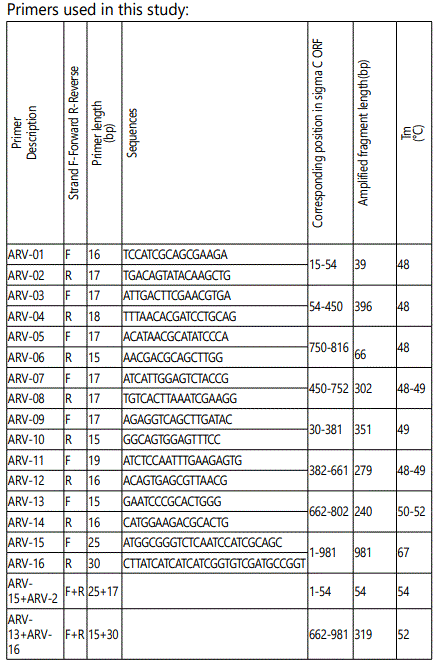
Viral RNA was extracted from the S1133 strain using a Trizol kit (Invitrogen, Carlsbad, CA). Synthesis of cDNA was performed as described (). The PCR product was electrophoresis in a 1% agarose gel and visualized by ethidium-bromide staining. The PCR product was purified using a Gene clean II kit (Qbiogene, Inc., Carlsbad, CA) and inserted into the 6.0 kb yeast expression vector, pNMT1-TOPO (Invitrogen, Carlsbad, CA). The ten pNMT1-σ C constructs was mobilized into competent yeast (Schizosachromycespombe) cells (Invitrogen, Carlsbad, CA), and transformed yeast cells were selected on plates containing Thiamine on EMM medium (Invitrogen, Carlsbad, CA).
The ten different size of sigma C proteins was expressed in an S. Pombe expression system (Invitrogen, Carlsbad, CA) and purified by Pro Bond purification system (Invitrogen, Carlsbad, CA) according to manufacturerʼs instructions. Briefly, transformed yeast containing thepNMT1-σ C or the control plasmid, pNMT1-TOPO without insert was used for further study. A single recombinant yeast colony containing the pNMT1-σ C construct was inoculated into 50 ml of EMM plus thiamine medium and grown overnight at 30°C with shaking. Cells were harvested by centrifugation at 1500x g for 5 min at room temperature. The supernatant was discarded. Cells were suspended in 50ml of EMM centrifuged, and the process was repeated. After final wash, cells were suspended in 50ml of EMM, and 500 µl aliquots of starter culture were inoculated into 100 ml of EMM and incubated at 30°C with shaking for 18 hr. Cells were harvested by centrifugation at1500 x g for 5 min at 4°C and washed in 10ml of 1 X TE buffer containing 100mM Nacl. Cells were centrifuged at 1500 x g for 5 min at 4°C and pellet suspended in 1 ml of 1 XTE + 100mM NaCl, and centrifuged for 2 min at top speed in the micro centrifuge. The supernatant was removed and 400µl of acid-washed glass beads added. Cells were broken apart using a Bead Beater (Scientific Industry Co., Bohemia, NY) at maximum speed for 45 seconds in a bead beater. Tubes were placed on ice for 5 min and this procedure repeated 5 times. Cells were centrifuged in a micro centrifuge for 2 min at maximum speed, and the supernatant was transferred to afresh tube. Protein concentration was determined by the Bradford method. The supernatant was further purified by a purification column (Invitrogen, Carlsbad, CA), briefly, add 8ml of supernatant to the column, bind for 30 mins, centrifuge (800xg) 5 mins, aspirate supernatant, wash with 8ml native wash buffer (provided by Invitrogen, Carlsbad, CA), centrifuge (800xg) 5mins, aspirate supernatant, repeat this process three more times, clamp the column in a vertical position and snap off the cap on the lower end. Elute the protein with 8ml native elution buffer (provided by Invitrogen, Carlsbad, CA), collect 1 ml fractions and analysis with SDS-PAGE and Western blot.
SDS-PAGE and western blotting
Western blot was performed as described previously. Proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane. The membrane was blocked with casein, and subsequently incubated for 1 hr with the chicken anti-reovirus sera that was diluted 1:100 in phosphate-buffered saline (PBS), washed three times with PBS, and incubated with rabbit anti-chicken immunoglobulin G (IgG) conjugated with horseradish peroxidase (Sigma, St. Louis, MO) diluted 1: 3000. After 1 hr of incubation at 37°C, the membrane was washed, and stained with Diaminonenzidine (Sigma, St. Louis, MO).

DNA sequencing and sequence analysis
To confirm the right clone of putative epitopes, 10 different fragments of the sigma C gene were cloned and sequenced.
Neutralization Assay
To detect the efficacy of different peptide against ARV, Neutralization assay was performed in Chicken Embryo Fibroblast cells (CEF). Briefly, CEFs were prepared from 9-day-old specific-pathogen-free (SPF) chicken embryonating eggs and grown in 96-well microplates. Chicken polyclonal antibody was used. Serial twofold dilutions were prepared of 150µl antibodies in 150µl minimum essential medium (MEM) containing antibiotics. Virus controls, negative serum control from uninfected chickens were also included. The S1133 virus dilution was added in 150µl amounts to all wells. The plates were incubated at 37°C in 5% CO2 for 1 hr. then 200µl of the different mixtures of virus and polyclonal antibody or virus, eptitope and polyclonal antibody was inoculated into CEF monolayers. After 60-min incubation at 37°C, inoculate were removed and monolayers were re-incubated with 1% FCS MEM. The neutralizing capacity was demonstrated by absence of CPE at 48 hr post-infection.
Results and Discussion
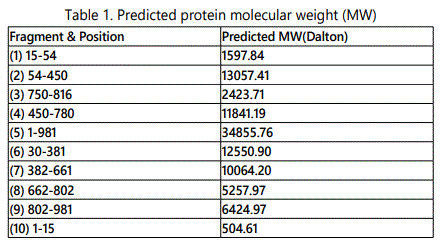
Expression and identification of sigma C proteins To create the full-length and deletion fragments of the sigma C gene of S1133 strain, PCR amplification using distinct primer pairs was carried out. Examination of all amplified PCR products following electrophoresis in agarose gels revealed the expected sizes. The resulting sigma C of the expression plasmid constructs was successfully over-expressed in S. pombe. Each plasmid DNA was introduced into S. pombe, and protein extracts of the induced recombinant cells were analyzed by 12% SDS-PAGE and could be visualized by Coomassie brilliant blue staining. Analysis of transformed and expression-induced yeast by SDS-PAGE revealed the expressed sigma C protein. The purified protein and protein extracts of the induced recombinant cells were then analyzed by Western blotting.
Virus-neutralization (VN)
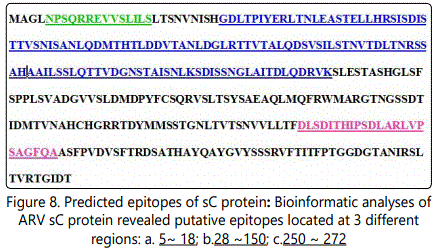
Bioinformaticsʼ analyses of ARV Sigma C protein revealed putative epitopes located at 3 different regions: a. 5 ~ 18; b. 28 ~150; c. 250 ~ 272test. Neutralization activity analysis of yeast expressed sigma C fragment revealed fragment 3, 6, 7, has the highest neutralization activity, which is corresponding to the position of sigma C protein of 750-816; 382-661 and 662-802. The actually result doesnʼt match with the prediction, it indicates current bioinformatics analysis we used doesnʼt fit for sigma C protein. Although our lab has produced two monoclonal antibodies against avian reovirus strain S1133, but no MAb had neutralizing activity against the tested reovirus, therefore, we are using polyclonal antibody in neutralization assay.
Acknowledgements
This research was supported by the National Science Foundationʼs Alliances for Graduate Education and the Professoriate (AGEP) Program, Grant No. 1432991, and USDA-IFAFS grant (#:00-52100-9705). We are grateful to Teresa Dormitorio and Chia-Chen Weng for their technical help.
References
- Spandidos DA, Graham AF. Complementation between temperaturesensitive and deletion mutants of reovirus. Journal of Virology. 1975; 16: 1444-1452.
- Glass SE, Naqi SA, Hall CF, Karr KM. Isolation and characterization of a virus associated with arthritis of chickens. Avian Dis. 1973; 17(2): 415-424. doi: 10.2307/1589226
- Bodelon G, Labrada L, Martinez-Costas J, Benavente J. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology. 2001; 290(2): 181-191. doi: 10.1006/viro.2001.1159
- Bodelon G, Labrada L, Martinez-Costas J, Benavente J. Modification of late membrane permeability in avian reovirus-infected cells: viroporin activity of the S1-encoded nonstructural p10 protein. J. Biol. Chem. 2002; 277(20): 17789-17796. doi: 10.1074/jbc.M202018200
- Robertson MD, Wilcox GE. Avian reovirus. Veterinary Bulletin. 1986; 56: 155-174.
- Wood GW, Nicholas RAJ, Hebert CN, Thornton DH. Serological comparisons of avian reoviruses. Journal of Comparative Pathology. 1980; 90(1): 29-38. doi: 10.1016/0021-9975(80)90025-0
- Hieronymus DR, Villegas P, Kleven SH. Identification and serological differentiation of several reovirus strains isolated from chickens with suspected malabsorption syndrome. Avian Dis. 1983; 27(1): 246-54. doi: 10.2307/1590390
- Wickramasinghe R, Meanger J, Enriquez CE, Wilcox GE. Avian reovirus proteins associated with neutralization of virus infectivity. Virology. 1993; 194(2): 688-96. doi: 10.1006/viro.1993.1309
- Varela R, Benavente J. Protein coding assignment of avian reovirus strain S1133. J. Virol. 1994; 68(10): 6775-6777.
- Liu HJ, Lee LH, Hsu HW, Kuo LC, Liao MH. Molecular evolution of avian reovirus: evidence for genetic diversity and reassortment of the S-class genome segments and multiple cocirculating lineages. Virology. 2003; 314: 336-349. doi: 10.1016/S0042-6822(03)00415-X
- Shapouri MR, Kane M, Letarte M, Bergeron J, Arella M, Silim A. Cloning, sequencing and expression of the S1 gene of avian reovirus. J Gen Virol. 1995; 76(6): 1515-20. doi: 10.1099/0022-1317-76-6-1515
- Wickramasinghe R, Meanger J, Enriquez CE, Wilcox GE. Avian reovirus proteins associated with neutralization of virus infectivity. Virology. 1993; 194(2): 688-96. doi: 10.1006/viro.1993.1309
- Yin. HS, H.K. Shieh, L.H. Lee. Characterization of the double stranded RNA genome segment S3 of avian reovirus. Journal of Virological Methods. 1997; 67(1): 93-101. doi: 10.1016/S0166-0934(97)00080-3
- Martinez-Costas J, Gonzalez-Lopez C, Vakharia VN, Benavente J. Possible Involvemnt of the Double-Standard RNA-Binding Core Protein ςA in the Resistance of Avian Reovirus to Interferon. J. Virol. 2000; 74(3): 1124-1131. doi: 10.1128/JVI.74.3.1124-1131.2000
- Chiu CJ, Lee LH. Cloning and nucleotide sequencing of the S4 genome segment of avian reovirus S1133. Arch. Virol. 1997; 142(12): 2515-2520.
- Yin HS, Lee LH. Identification and characterization of RNA-binding activities of avian reovirus non-structural protein σNS. J. Gen. Virol. 1998; 79: 1411-1413. doi: 10.1099/0022-1317-79-6-1411
- Huang PH, Li YJ, Su YP, Lee LH, Liu HJ. Epitope mapping and functional analysis of sigma A and sigma NS proteins of avian reovirus. Virology. 2005; 332 (2): 584-595. doi: 10.1016/j.virol.2004.12.005


