Research Article
Low pathogenic Avian Influenza Virus circulating in wild Aquatic Birds of Southern US
Department of Biological Sciences, Alabama State University, Montgomery, AL 36101 USA
*Corresponding author: Hongzhuan Wu, Professor, Department of Biological Sciences, Alabama State University, AL 36101, USA, E-mail: hwu@alasu.edu
Received: October 13, 2017 Accepted: October 26, 2017 Published: November 2, 2017
Citation: Bolden KE, Austin J, Scissum-Gunn K, Robertson BK, Villafane R, Wu H. Low pathogenic Avian Influenza Virus circulating in wild Aquatic Birds of Southern US. Madridge J Clin Res. 2017; 2(1): 31-36. doi: 10.18689/mjcr-1000106
Copyright: © 2017 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Wild aquatic birds, predominantly duck, geese and shorebirds, are natural reservoir of all subtypes of influenza A viruses. Avian influenza viruses are classified into two pathogenic types: low pathogenic avian influenza (LPAI) virus and highly pathogenic avian influenza (HPAI) virus. To date, LPAI has been isolated from migratory waterfowls, and the adaption of these viruses to land fowls might lead to the generation of HPAI which is a public health concern. The avian influenza virus is shed in large amounts in the feces, and can spread between birds by the fecaloral route and cause asymptomatic or low pathogenic infection. Alabama and Georgia are located in the flyways of wild migrating birds during the winter season, AIV surveillance of the samples from these states will be of great interest for the study of the epidemiology of LPAI virus. In the present study we analyzed the prevalence of AIV circulating in Alabama and Georgia, Real time RT-PCR was performed on 360 fecal samples. An overall AIV prevalence of 5% was detected with a peak during the spring and winter; resulting PCR product was further confirmed by sequencing, in which H1N1 subtype was identified. Our results support fecal sampling as an appropriate method for large-scale LPAI virus surveillance programs.
Keywords: Avian influenza virus; Aquatic birds; Fecal samples; qPCR.
Abbreviations: AGID: Agar Gel Immunodiffusion; AI: Avian Influenza; AIV: Avian Influenza Virus; HA: Hemagglutinin; HPAI: Highly Pathogenic Avian Influenza; IVPI: Intravenous Pathogenicity Index; LP: Low Pathogenic; LPAI: Low Pathogenic Avian Influenza; LPAIV: Low Pathogenic Avian Influenza Virus; M: Matrix Protein; NA: Neuraminidase; NP: Nucleoprotein; PBS: Phosphate Buffered Saline; RRT-PCR: Real-time Reverse Transcription PCR; RTPCR: Reverse Transcriptase Polymerase Chain Reaction.
Introduction
Avian influenza (AI) is caused by viruses that are members of the family Orthomyxoviridae [1]. The influenza viruses that make up this family are classified into three types; A, B, or C based upon the differences between their nucleoprotein and matrix protein antigens [2]. Only influenza A viruses are known to infect birds. Influenza viruses are further categorized into subtypes according to antigens of the hemagglutinin and neuraminidase projections on their surfaces. Influenza A viruses have been represented by all subtypes of hemagglutinin and neuraminidase. Avian influenza viruses (AIV) cause disease in poultry at varying levels of severity according to the strain of virus involved. Strains are classified into low and highly pathogenic types depending on the severity of diseases in which they cause [3, 4]. However, there is a possibility that viruses may move between low and highly pathogenic due to mutations or genetic re-assortments.
Diagnosis of avian influenza viruses is done by isolation and characterization of the virus. Infections in birds can cause numerous varieties of clinical signs that may vary according to the host, strain of virus, the hostʼs immune system, existence of secondary exacerbating organisms, and environmental conditions [5]. Diagnosis of these viruses is important because infections in birds can cause serious illness, and even lead to death.
Domestic fowl, ducks, geese, turkeys, quail, and other avian species are all susceptible to contracting avian influenza. Disease outbreaks occur in domestic fowl and turkeys [5]. Numerous species of wild birds, particularly water birds are most susceptible to contracting avian influenza. Pathogenic strains of AIV can appear at any time and cause disease in domestic poultry in any country at any time. There have been outbreaks that occurred on all continents. The most severe outbreaks have been reported in Hong Kong, Chile, The Netherlands and South East Asia.
There is need to prevent the spread of the avian H1N1 virus to swine or humans to prevent viral genome recombination. This is currently done using quarantine measures, culling of infected poultry flocks, swine farms, and improved biosecurity. However, vaccination of humans and swine remains an important tool to prevent spread between species. The prerequisite for controlling the disease is rapid and accurate identification of this virus. The agar gel immunodiffusion (AGID) test remains the diagnostic assay most utilized for detection of AIV antibodies in commercial poultry worldwide and is considered the “gold standard” by the World Organization for Animal Health [6]. The AGID test detects antibodies to two influenza virus proteins, NP and M1, which are highly conserved and type specific. This assay is time consuming and lacks an ability to distinguish amongst AIV subtypes. Reverse transcriptase polymerase chain reaction (RT-PCR) and real time RT-PCR (RRT-PCR) has been developed as rapid detection tests for AIVs [7, 8]. Our previous research proposed a laboratory protocol to detect AIV from wild aquatic birds; this continuation research successfully detected AIV samples from Alabama and Georgia and revealed molecular epidemiology of LP AIV in part of the southern states.
In order to implement effective control measures there must be epidemiological studies, and the massive amount of wild migratory birds that land in Alabama and Georgia provides an appropriate model situation in which to examine the epidemiological relationship, which influences transmission of low pathogenic avian influenza viruses.
Because most wild bird species considered to be avian influenza virus reservoirs are migratory, they are suspected of having a role in the spread of AIV. Avian influenza usually involves migratory waterfowl. It has been proved that highly pathogenic avian influenza viruses surface in domestic poultry from low pathogenic progenitors of the H5 and H7 subtypes. There has been increased virus surveillance and establishment of AIV databases, not only in the United States of America, but also around the world.
The term avian influenza refers to influenza A viruses, which is predominantly found in birds. However, infections with this particular virus can be found in humans. The risk of human infection by the avian influenza virus is normally low to most people because this virus does not usually infect humans. However, there have been reports of confirmed cases of human infection from numerous subtypes of avian influenza since 1997. Most of these seen cases of human infection resulted from contact with infected poultry. The increase in the occurrence of avian influenza worldwide in poultry and humans introduces the potential for another influenza A pandemic that could pose a threat to both human health and the global economy [9].
The past pandemic strain of H1N1 has now become a common component of current seasonal influenza viruses [10]. The reoccurring of this virus has changed the pre-existing immunity of the human population to succeeding infections. The numerous circulating subtypes of avian influenza virus propose the threat of the virus re-assorting into new forms [11, 12]. Identifying genetic mutations and antigenic properties would be useful in developing vaccines and aiding in the control of influenza.
The study and detection of low pathogenic avian influenza helps fill in important gaps in the understanding of the global movement of avian influenza viruses, the reservoir species they are found in, and their prospective impact on animal and human health. By understanding the epidemiology of avian influenza it aids in forming and implementing control policies for highly pathogenic avian influenza. Examination of avian influenza is highly important because there have been several bird flu outbreaks since the first one in December of 2003 [13]. Wild birds as well as farmed livestock have died due to bird flu in Africa, Asia and Europe. Human cases were also reported and were related to the strains of H5 and H7 [14-20].
Avian influenza is a serious transmissible disease of all birds. AI has been diagnosed in several states of the United States [21-28]. Any form of outbreak of avian influenza in Alabama or Georgia could put at risk the entire poultry industry of these two states. In this study, we are aiming to investigate the LPAIV epidemiology in southern US by detecting HA gene of RT-qPCR, and compare temperature, sampling season and sampling treatment essay to the effect of detection, also by using sequencing and phylogenetic tree analysis to reveal the virus relationship with the previous outbreak strain.
Materials and Methods
Sampling
Most sampling was conducted in late September to early February, from 2013 to 2016. Samples were directly from fecal samples nesting waterfowl (Figures 3 & 4) from parks in Alabama and Georgia (Figure 1). Swabs were placed in a tube containing 2 mL of virus-transport medium (PBS plus 5% fetal bovine serum, supplemented with 10,000IU/mL of penicillin G and 10 g/mL of streptomycin Sulfate), kept cold in the field with wet ice or frozen in dry ice and transferred to a −70°C freezer within 5h. Frozen tubes containing cloacal swabs in transport medium were thawed and centrifuged at 2,000 × g for 10 min. The cotton-tipped swab was discarded and the supernatant was aliquoted into 3 vials for use in egg embryo inoculation (aliquot 1), and RRT-PCR (aliquot 2) as shown in Figure 2. Control AIV virus H10N7 strain was provided by Dr. Giambroneʼs lab.
Experimental Design Outline
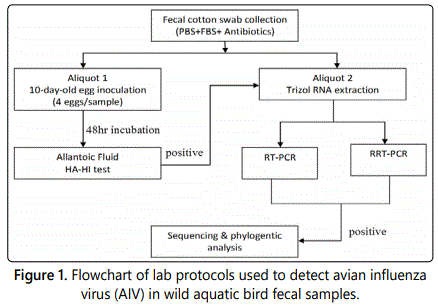
Real-Time Reverse Transcription PCR (RRT-PCR)
Total RNA was extracted from fecal samples using TRIzol (Invitrogen, Carlsbad, CA) as described in the manual. Five micrograms of total RNA were treated with 1.0 unit of DNAse I and 1.0µl of 10 x reaction buffer (Sigma, St. Louis, MO), incubated for 15 min at room temperature, 1.0 µl of stop solution was added to inactivated DNase I, and the mixture was heated at 70 0C for 10 min. RNA was reverse transcribed using Superscript first-strand synthesis system (Invitrogen, Carlsbad, CA) according to manufacturerʼs recommendations. Oligonucleotide primers sequences, forward primer Bm-HA-(TATTCGTCTCAGGGAGCAAAAGCAGGGG) and reverse primer Bm-NS 890R (ATATCGTCTCGTATTAGTAGAAACAAGGGTGTTTT). Amplification and detection were carried out using equivalent amounts of total RNA from samples using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA). The relative transcriptional levels of virus RNA were determined by subtracting the cycle threshold (Ct) of the sample by that of the GAPDH, the calibrator or internal control, as per the formula: DCt = Ct (sample) _ Ct (calibrator). The relative virus RNA copy compared to that in negative control was then calculated by the formula 2DDCt where DDCt =DCt (sample) _DCt (negative control).
The RRT-PCR results are reported as positive or negative (amplification or no amplification) for the target gene, with a corresponding Cycle threshold (Ct)value, which represented the point at which amplification of DNA is detected above background fluorescence (ABI Instrument Operatorʼs Manual). A sample with a lower initial concentration of target DNA required more cycles to reach the Ct. Samples with a higher concentration required fewer cycles.
Sequencing and Phylogenetic Analysis
Purification of the PCR product was done by using the Wizard PCR Preps DNA Purification System from Promega. The PCR product was prepared by quickly excising the DNA band from the gel. The agarose slice was then transferred to a 1.5ml microcentrifuge tube, and 1ml of resin was added. The gel was then incubated at 650 C for 5 minutes to allow the gel to melt. The PCR product was then purified by preparing a wizard column for each purified sample, a Syringe Barrel was attach to the Minicolumn. The resin/DNA mix was added to the syringe barrel, and a plunger was inserted in order to push the resin/ DNA slurry into the Minicolumn. Adding 2ml of 80% isopropanol washed the PCR product, the isopropanol was pushed through the Minicolumn using a plunger. The Minicolumn was then transferred to a microcentrifuge tube and centrifuged at 10,000 x g for 2 minutes. By transferring the Minicolumn to a clean 1.5ml microcentrifuge tube, it eluted the PCR. 50ul of Nuclease-Free Water was added to the tube. The mixture was then centrifuged at 10,000 x g for 20 seconds at room temperature. Finally, the Minicolumn was removed and discarded, and stored at -200 C until sent off to Auburn University for sequencing. The ABI PRiSM Genetic Analyzer at Auburn University used the purified PCR product as the template during automated sequencing. The sequence provided was then compared to previously published HA subtype sequences to determine the genotype. A phylogenetic tree between HA subtypes was also generated with NCBI by the neighbor-joining method.
Results
Detection of field sample with RT-qPCR
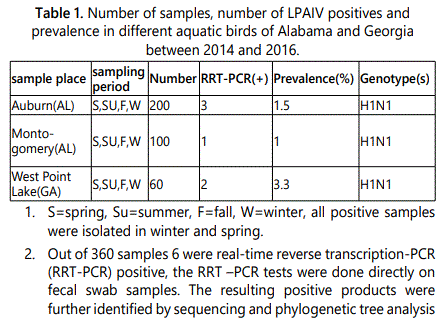
Real-time reverse transcription PCR was performed on all 360 samples. The results from the real-time reverse transcription PCR indicated that 6 out of the 360 samples were positive for the avian influenza virus of wild aquatic birds in Alabama and Georgia (Figure 2). Out of the 200 samples collected from Auburn, Alabama, 3 were indicated as positive giving 1.5% prevalence. From the 100 samples taken from Montgomery, Alabama, 1 was indicated as positive giving 1% prevalence. The 60 collected samples from West Point Lake, Georgia resulted in 2 positives giving 3.3% prevalence.
Temperature effect to positive rate
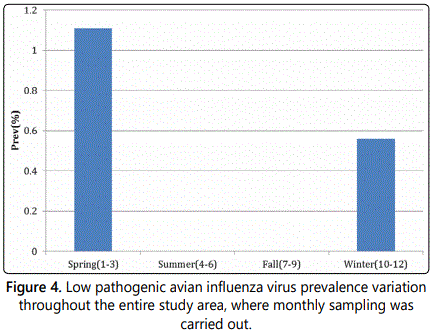
The mean seasonal temperature was recorded for all seasons throughout the sampling area (Figure 3). The seasons of spring and winter had the lowest temperatures compared to that of the summer and fall seasons. The low temperatures of spring and winter contributed to the positive prevalence of avian influenza in the research study (Figure 4). The positive results seen during the spring and the winter indicates that the lower temperatures of these two seasons preserved the virus in the fecal samples longer than that of the summer and the fall. The heat of the summer and fall inactivated the virus that was present in the fecal samples.

Freshness of sample to the positive rate
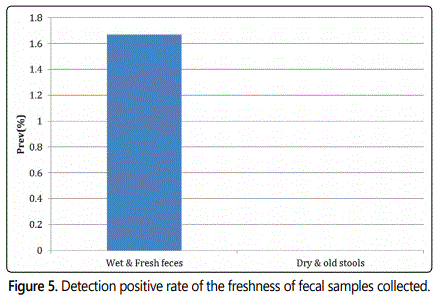
Fresh feces and old feces were collected from the collection sites in Alabama and Georgia. The freshness of the fecal samples collected were compared, it was found that the fresh feces resulted in the positive prevalence of avian influenza (Figure 5).
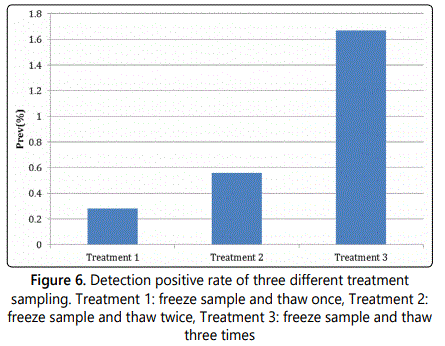
Sample treatment to the positive rate
Also, treatments were performed on the fecal samples. There were three different treatment methods performed on the samples, which included: Treatment 1: freezing the sample and thawing it one time, Treatment 2: freezing the sample and thawing it two times, and Treatment 3: freezing the sample and thawing it three times (Figure 6). Using treatment method 3 gave the highest detection positive rate for avian influenza. Freezing and thawing the samples allowed the virus to be released from the cell and excreted into the transport medium.
Phylogenetic tree analysis to the positive samples
Once it was confirmed that avian influenza was detected in the collected samples, sequencing was done from the purified PCR product (Figure 7), and a phylogenetic tree analysis was constructed. Out of all 360 samples that were collected, the 6 that resulted in being positive were all classified as H1N1 low pathogenic avian influenza virus subtype (Table 1). There was an overall LPAIV prevalence of 1.67% collected from the wild birds of these two states.
The results from the phylogenetic tree analysis shows that the H1N1 subtype that was collected from the states of Alabama and Georgia was in the same clade as the H1N1 subtypes isolate in 2008 from Alabama (A/Alabama/AF1952/2008(H1N1) (Figure 8). This indicates that there is little to no genetic differences. The two have the same genotype, which means that there have been no mutations. The isolates that were obtained from the spring and winter seasons are still the same as the older strain from 2008. There were other viruses found, although not in the same clade, they still belonged to the H1N1 subtype.
Discussion
Understanding the epidemiology of avian influenza viruses has been regarded as imperative in devising and executing control policies for highly pathogenic avian influenza. Although avian influenza has been recognized for a substantial amount of time, its pattern of occurrence has still amazed scientists around the world due to its ever-changing appearance.
The study of avian influenza epidemiology helps scientists understand how and why highly pathogenic avian influenza viruses emerge from low pathogenic avian influenza viruses. The examination of ecology and evolution of AIVs in the natural host is key to understanding the role that wild birds might play in circulating viruses among different geographic regions [29].
The prevalence of 1.67% was not very high, which means that not all migratory avian species carry the influenza virus. The low prevalence could also be due to the climate and season in which the samples were collected, both which influences the detection of avian influenza virus. It appeared that the lower the temperatures were, the longer the virus survived in the fecal samples. The cooler temperatures gave the highest positive prevalence for avian influenza. Collecting more fecal samples from numerous geographic areas of the states of Alabama and Georgia can possibly increase the prevalence for avian influenza in each city of the two states.
In addition, by identifying the H1N1 subtype it allows the human population to know which strand of avian influenza virus is circulating in aquatic birds of Alabama and Georgia at this present time. By knowing the strand of the circulating virus, it allows researchers to evaluate how close a match there is between viruses used to make the vaccine and circulating viruses. Traditional flu vaccines are constructed to protect against influenza A (H1N1) virus, influenza A (H3N2) virus, and influenza B virus [30-35].
The changes that are seen in climatic conditions, host population dynamics, and host population immunity are possibly substantial to understanding the epidemiology of avian influenza virusʼ natural host(s) [36]. Gaining more knowledge about such factors will require information about infection from ecologically connected populations over longer intervals of time, and across larger dimensions when studying migratory birds.
For future work, more fecal samples should be collected from additional southern states. The fecal samples should be collected over a span of years and varying seasons. It would be of great interest to see if there is a significant change in the prevalence of avian influenza virus, and also a change in the subtype of the identified avian virus. Also, the sampling and treatment process should be further optimized. The third freeze/thaw treatment process should be added to the initial protocol because it was the most effective process. It is possible that false negatives resulted from only doing the freeze/thaw process only once. The type of avian feces collected should be taken into consideration, seeing that the fresh feces in this research study gave the only positive prevalence of avian influenza. The weather conditions should be taken into consideration before collecting samples. It is possible that the feces that appeared wet and fresh were wet by rainfall, which interfered with the positive prevalence detection for avian influenza in this study. In addition, the time it takes to get the samples from the collection site back to the lab should also be taken into consideration. It is possible that the virus is capable of being inactivated during transport, resulting in a low detection prevalence.
Capturing the avian species then collecting the sample represents previous protocol for the detection of avian influenza. However, that method is very time consuming. Being aware that avian influenza virus is shed in large amounts in the feces set the standard by which this research was performed. The procedures used in this research to detect avian influenza viruses directly from fecal samples are a more convenient method than previous reported cloacal swabs protocol.
In conclusion our research reveals we still have the old AIV strain circulate in southern US, recent outbreak of H7N9 in Asia hasnʼt been brought into here by migratory birds, H7N9 has been proved could be spread among mammals, with winter season coming, we need to monitor AIV more frequently to avoid highly pathogenic AIV into US through birds which can cause pandemic in human.
Acknowledgements
This work was supported by NSF-REU (DBI-1358923) to Dr. KomalVig (PI) and by NSFCREST (HRD-1241701) to Dr. Shree S. Singh (PI).
References
- Causey D, Edwards SV. Ecology of Avian Influenza Virus in Birds. The Journal of Infectious Diseases. 2008; 197(1): 29-33. doi: 10.1086/524991
- Weis W, Brown JH, Cusack S, et al. Structure of the Influenza virus hemagglutinin complex with its receptor, sialic acid. Nature. 1988; 333(6172): 426-31. doi: 10.1038/333426a0
- Piaggio AJ, Shriner SA, VanDalen KK, et al. Molecular Surveillance of Low Pathogenic Avian Influenza Viruses in Wild Birds across the United States: Inferences from the Hemagglutinin Gene. PLoS ONE. 2012. doi: 10.1371/journal.pone.0050834
- Vergara-Alert J, Moreno A, Zabala GJ, et al. Exposure to a Low Pathogenic A/H7N2 Virus in Chickens Protects against Highly Pathogenic A/H7N1 Virus but Not against Subsequent Infection with A/H5N1. PLoS ONE. 2013. doi: 10.1371/journal.pone.0058692
- Watson DS, Reddy SM, Brahmakshatriya V, Lupiani B. A Multiplexed Immunoassay for Detection of Antibodies against Avian Influenza Virus. Journal of Immunological Methods. 2009; 340(2): 123-31. doi: 10.1016/j.jim.2008.10.007
- Ellis JS, Zambon MC. Molecular Diagnosis of Influenza. Reviews in Medical Virology. 2002; 12(6): 375-89. doi: 10.1002/rmv.370
- Kok J, Blyth CC, Foo H, et al. Comparison of a Rapid Antigen Test with Nucleic Acid Testing during Cocirculation of Pandemic Influenza A/H1N1 2009 and Seasonal Influenza A/H3N2. Journal of Clinical Microbiology. 2010; 48(1): 290-291. doi: 10.1128/JCM.01465-09
- Whitley RJ, Monto AS. Seasonal and Pandemic Influenza Preparedness: A Global Threat. The Journal of Infectious Diseases. 2006; 194(2): 65-69. doi: 10.1086/507562
- Lara-Ramírez EE, Segura-Cabrera A, Salazar MI, Rodriguez-Perez MA, Guo X. Large Scale Genome Analysis Shows That the Epitopes for Broadly CrossReactive Antibodies Are Predominant in the Pandemic 2009 Influenza Virus A H1N1 Strain. Viruses. 2013; 5(11): 2796-802. doi: 10.3390/v5112796
- Neumann G, Chen H, Gao GF, Shu Y, Kawaoka Y. H5N1 Influenza Viruses: Outbreaks and Biological Properties. Cell Research. 2009; 20: 51-61. doi: 10.1038/cr.2009.124
- Cao S, Liu X, Yu M, et al. A Nuclear Export Signal in the Matrix Protein of Influenza A Virus Is Required for Efficient Virus Replication. Journal of Virology. 2012; 86(9): 4883-891. doi: 10.1128/JVI.06586-11
- Ely CR, Hall J, Schmutz JA, et al. Evidence That Life History Characteristics of Wild Birds Influence Infection and Exposure to Influenza A Viruses. PLoS ONE. 2013; 8(3). doi: 10.1371/journal.pone.0057614
- Costa TP, Brown JD, Howerth EW, Stallknecht DE. The Effect of Age on Avian Influenza Viral Shedding in Mallards (AnasPlatyrhynchos). Avian Diseases. 2010; 54(1): 581-85. doi: 10.1637/8692-031309-ResNote.1
- Cox NJ, Subbarao K. Global Epidemiology of Influenza: Past and Present. Annual Review of Medicine. 2000; 51: 407-21. doi: 10.1146/annurev. med.51.1.407
- Horimoto T, Kawaoka Y. Pandemic Threat Posed by Avian Influenza A Viruses. Clinical Microbiology Reviews. 2001; 14(1): 129-49. doi: 10.1128/CMR.14.1.129-149.2001
- Alexander DJ, Capua I. Avian influenza infection in birds: A challenge and opportunity for the poultry veterinarian. Poultry Science. 2009; 88(4): 842-846. doi: 10.3382/ps.2008-00289
- Whitley RJ, Monto AS. Seasonal and Pandemic Influenza Preparedness: A Global Threat. The Journal of Infectious Diseases. 2006; 194(2): 65-69. doi: 10.1086/507562
- Mumford E, Bishop J, Hendrickx S, Embarek PB, Perdue M. Avian influenza H5N1: Risks at the human-animal interface. Food Nutr Bull. 2007; 28(2): 357-363. doi: 10.1177/15648265070282S215
- Huang S, Chen C, Hu Y, et al. Overview on Prevention and Control Measures for Human Infection with HPAI H5N2 Virus in Taiwan in 2012. Epidemiology Bulleten. 2013; 29(17): 203-204. doi: 10.6525/TEB
- Iskander J, Strikas RA, Gensheimer KF, Cox NJ, Redd SC. Pandemic Influenza Planning, United States, 1978–2008. Emerging Infectious Diseases. 2013; 19(6): 879-85. doi: 10.3201/eid1906.121478
- Anhlan D, Grundmann N, Makalowski W, Ludwig S, Scholtissek C. Origin of the 1918 Pandemic H1N1 Influenza A Virus as Studied by Codon Usage Patterns and Phylogenetic Analysis. RNA. 2010; 7(1): 64-73. doi: 10.1261/rna.2395211
- Aamir U, Badar N, Rashid M, et al. Molecular Epidemiology of Influenza A(H1N1)pdm09 Viruses from Pakistan in 2009–2010. PLoS ONE. 2012; 7(8). doi: 10.1371/journal.pone.0041866
- Lewis N, Javakhishvili Z, Russell C, et al. Avian Influenza Virus Surveillance in Wild Birds in Georgia: 2009–2011. PLoS ONE. 2013; 8(3). doi: 10.1371/journal.pone.0058534
- Peng Y, Zhi-Xun X, Jia-Bo L, et al. Epidemiological Surveillance of Low Pathogenic Avian Influenza Virus (LPAIV) from Poultry in Guangxi Province, Southern China. PLoS ONE. 2013; 8(10). doi: 10.1371/journal.pone.0077132
- Li Q, Zhao Q, Gu M, et al. Genome Sequence of a Novel Reassortant H3N2 Avian Influenza Virus from Domestic Mallard Ducks in Eastern China. Genome Announcements. 2013; 1(2). doi: 10.1128/genomeA.00221-12
- Suarez D, Senne DA, Banks J, et al. Recombination Resulting in Virulence Shift in Avian Influenza Outbreak, Chile. Emerging Infectious Diseases. 2004; 10(4): 693-99. doi: 10.3201/eid1004.030396
- Iskander J, Strikas R, Gensheimer K, Cox NJ, Redd SC. Pandemic Influenza Planning, United States, 1978–2008. Emerging Infectious Diseases. 2013; 19(6): 879-885. doi: 10.3201/eid1906.121478
- Tang RB, Chen HL. An Overview of the Recent Outbreaks of the Avianorigin Influenza A (H7N9) Virus in the Human. Journal of the Chinese Medical Association. 2013; 76(5): 245-48. doi: 10.1016/j.jcma.2013.04.003
- Carver S, Kilpatrick A, Kuenzi A, et al. Environmental monitoring to enhance comprehension and control of infectious diseases. Journal Of Environmental Monitoring. 2010; 12(11): 2048-2055. doi: 10.1039/c0em00046a


