Research Article
Metastasis-Associated Lung Adenocarcinoma Transcript 1 Expression Levels in Egyptian Patients with Multiple Myeloma: Relation to Disease Characteristics and Possible Prognostic Implication
Department of Hematology, Medical Research Institute, Alexandria University, Egypt
*Corresponding author: Maha M. A. El Gammal, Department of Hematology, Medical Research Institute, Alexandria University, Egypt, E-mail: mahagammal@yahoo.com
Received: July 10, 2021 Accepted: August 05, 2021 Published: August 10, 2021
Citation: El Gammal MMA, Sultan MM, Sultan HK, Balbaa OA, Shaaban IOA. Metastasis-Associated Lung Adenocarcinoma Transcript 1 Expression Levels in Egyptian Patients with Multiple Myeloma: Relation to Disease Characteristics and Possible Prognostic Implication. Madridge J Cancer Stud Res. 2019; 4(1): 97-102. doi: 10.18689/mjcsr-1000114
Copyright: © 2021 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Multiple myeloma (MM) is a malignancy of antibody-secreting plasma cells which remains incurable, despite significant improvements in treatment and patient care. There is an urgent need to identify novel markers with prognostic and therapeutic value for MM. Long non-coding RNAs (lncRNAs) have emerged as key regulators in cancers including MM. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA known to be over expressed in solid tumors and hematologic malignancies. However, the pathological mechanisms of MALAT1 in MM are not completely understood.
Objective: To investigate the MALAT1 expression levels in MM patients and to evaluate their relations to disease characteristics and possible prognostic implication.
Patients and methods: The study was performed on 50 MM patients and 50 patients doing bone marrow aspiration for other conditions with normal plasma cell percentage as controls. Total RNA, including LncRNA isolation from bone marrow samples was carried out with the miRN easy Mini Kit according to the manufacturer’s instructions. MALAT1 gene expression was performed by RNA extraction, reverse transcription, and real-time qPCR.
Results: We demonstrated that MALAT1 was over expressed in newly diagnosed MM patients compared with post-treatment patients and control subjects. Additionally, there were statistically significant relations between high MALAT1 expression levels and ESR, plasma cell percentages in bone marrow, M-protein concentrations in serum protein electrophoresis (SPEP), hemoglobin levels and serum levels of β2-microglobulin and IL-6.
Conclusion: The current data suggested that MALAT1 may be useful as a novel prognostic biomarker for MM.
Keywords: MALAT1, Multiple Myeloma, lncRNAs
Introduction
Multiple myeloma (MM) is characterized by the malignant proliferation of plasma cells in the bone marrow microenvironment, accompanied by organ dysfunctions, including anemia, renal insufficiency, hypercalcemia, and osteolytic bone disease [1]. It is the second commonest hematological malignancy with rising incidence across the globe [2].
MM is still an incurable disease, despite advances in conventional therapy, clinical application of novel agents and high-dose chemotherapy protocols supported by autologous stem cell transplantation [3]. Thus, a detailed understanding of the mechanisms underlying the development and progression of MM is essential for improving the treatment. Growing evidence indicates the involvement of lncRNAs in MM pathogenesis, providing new insights into its biological mechanisms [4].
LncRNAs, with a length of over 200 nucleotides, have little or no capacity for protein synthesis. These have been shown to regulate gene expression at the transcriptional, posttranscriptional, and epigenetic levels [5] and are implicated in diverse biological functions, including development, proliferation, differentiation, and apoptosis [6]. The expressions and roles of lncRNAs in various benign and malignant diseases have been widely investigated [7–9]. Aberrant expression of lncRNAs has been observed in many types of hematological malignancies, suggesting that they own either oncogenic or tumor suppressive properties [10]. In addition to crucial role in tumor development and progression, lncRNAs showed many interesting features as diagnostic and predictive biomarkers and also as promising therapeutic targets [11].However, the available data suggest important role of lncRNAs in MM [12].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a 8.5-Kb located on chromosome 11 (11q13.1); a translocation site of t(11; 14) in MM, consists of two exons and is one of the few biologically well-studied lncRNAs, and it has been reported to involve the regulation of gene expressions and the regulation of alternative splicing and cell cycle [13]. MALAT1 misregulation has been shown to play a role in the development of several solid tumors, including lung, colorectal, bladder, and laryngeal cancers [14-16].
Nevertheless, the impact of MALAT1 on MM has not yet been satisfactorily investigated. To understand the potential role of MALAT1 in MM, we tested the expression level of MALAT1 in three groups of MM patients and analyzed its relationship to different patients’ characteristics and prognosis.
Participants and methods
Participants: This study was conducted in the period between January 2018 and December 2018 and included 100 subjects that were classified into 2 groups. The first group included 50 MM patients, who were diagnosed according to the international myeloma working group (IMWG) criteria for the diagnosis of MM, which was subdivided into three subgroups: T0 group consisting of 12 newly diagnosed multiple myeloma patients who had received no treatment at the time of collection of samples, T1 group consisting of 27 MM patients who have received 2 cycles of velcade and dexamethasone or velcade, cyclophosphamide and dexamethasone at the time of collection of samples and T2 group consisting of 11 patients who have received 4 cycles of velcade and dexamethasone or velcade, cyclophosphamide and dexamethasone at the time of collection of samples. While the second group included 50 patients doing bone marrow (BM) aspiration for other conditions (as hypersplenism or primary immune thrombocytopenia) whose BMs showed normal plasma cell count (less than 3% plasma cells). All patients were recruited from the Hematology Department in Gamal Abdel Nasr Hospital; Alexandria affiliated to Health Insurance organization.
All subjects included in the current study were subjected to full medical checkup as history taking, clinical examination, laboratory investigations including BM examination, IL-6 measurement, radiological examination and MALAT1 expression by quantitative real-time PCR assays.
MM patient’s group was assessed at three time points: Time 0 was the time of diagnosis where the patients have had no treatment, Time 1 was the time at which the patients had received 2 cycles of velcade and dexamethasone or velcade, cyclophosphamide and dexamethasone at the time of collection of samples and Time 2 was the time at which the patients had received 4 cycles of velcade and dexamethasone or velcade, cyclophosphamide and dexamethasone at the time of collection of samples. Whereas, the assessment of the laboratory parameters in the control group was done only once at the time of collection of samples.
The updated criteria of International Myeloma Working Group (IMWG) were used as a guide for proper diagnosis of MM patients [17]. The treatment response categories were in accordance with the uniform response criteria created by IMWG [18].
This study was approved by the local ethics committee at the Medical Research Institute, Alexandria University, Egypt. Informed consent from the patients was obtained before sample collection and after a brief explanation of research objectives.
Measurement of serum IL-6 levels: IL-6 level was measured using a Cobas E601 analyzer (Roche Diagnostics) for all subjects included in the current study [19].
MALAT1 gene expression by real-time PCR
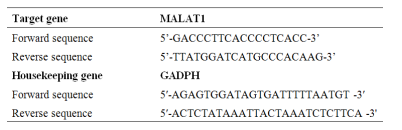
RNA extraction was performed at Hematology Department Medical Research Institute, Alexandria University via Qiagen Blood QIAamp genomic RNA extraction kit and under sterile conditions using a UV laminar flow cabinet according to manufacturer instructions. The quality and quantity of RNA samples were analyzed and controlled at the end of extraction by NanoDrop 2000 spectrophotometer (Thermo Scientific). Reverse transcription (RT) was performed using QuantiTect Reverse Transcription Kit (Qiagen). The quality and quantity of DNA samples were analyzed and controlled at the end of RT by NanoDrop 2000 spectrophotometer. SYBR Green real-time quantitative PCR preparation was performed under sterile conditions using a UV laminar flow and the reaction was performed using RotorGene Q cycler from Qiagen. Primers details are supplied in table I [20]. Ct values were evaluated for cases and controls, ΔΔCt values (ΔCt case - ΔCt control), and the relative gene expression was calculated using 2-ΔΔCt[21].
Results:
Demographic characteristics of the MM groups were shown in Table II.
Expression levels of MALAT1 in MM patients and healthy individuals:
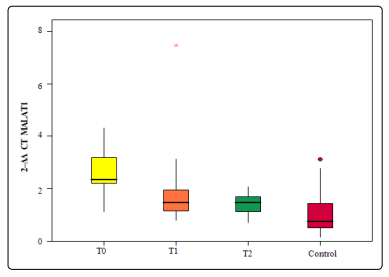
MALAT1 expression levels were significantly higher at diagnosis (T0) as well as after 2 cycles of bortezomib-based therapy (T1) compared with those levels reported after 4 cycles of bortezomib containing therapy (T2) in MM patients or control subjects. However, the differences were not statistically significant between the expression levels reported in T1 when compared to T2 MM patients as illustrated in figure 1.
Clinical importance of dysregulated MALAT1 in MM:
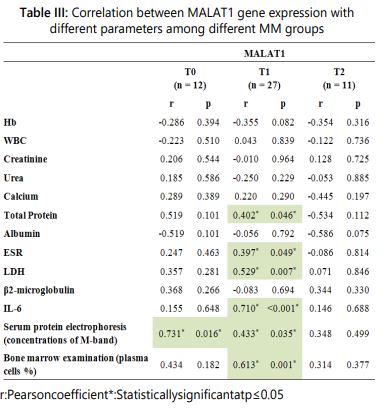
The associations between the MALAT1 expression and different aspects of disease were further analyzed. In the T0 group, the MALAT1 gene expression levels showed a statistically significant positive correlation only with M-protein concentration in SPEP but not to other disease parameters. The MALAT1 gene expression levels in the T1 group showed significant positive correlations with PC percentages in BM, M-protein concentration in SPEP, ESR, and serum levels of LDH, total protein, as well as IL-6. Lastly, its expression levels in the T2 group displayed no significant correlation to all disease parameters studied as shown in table III. These findings show that MALAT1 may be useful as a novel prognostic biomarker for MM.
Furthermore, it is noteworthy to mention that in the current study two MM cases presented with extra- medullary infiltration. One case died shortly after diagnosis. Both cases had Stage III. They had high concentrations of M-protein in SPEP and both were of IgG kappa type. Both cases presented with CRAB criteria and poor prognostic features in the form of high concentrations of β2- microglobulin and IL-6 levels and low albumin levels. The percentages of PCs in the BM at diagnosis of both cases were very high (>80%). The MALAT1gene expression levels in both cases were high. Correlations between MALAT1 gene expression and extramedullary myeloma spread were not applicable as only two cases showed extramedullary spread (EMM).
Relation between different levels of MALAT1 gene expression (high – low) with other parameters:
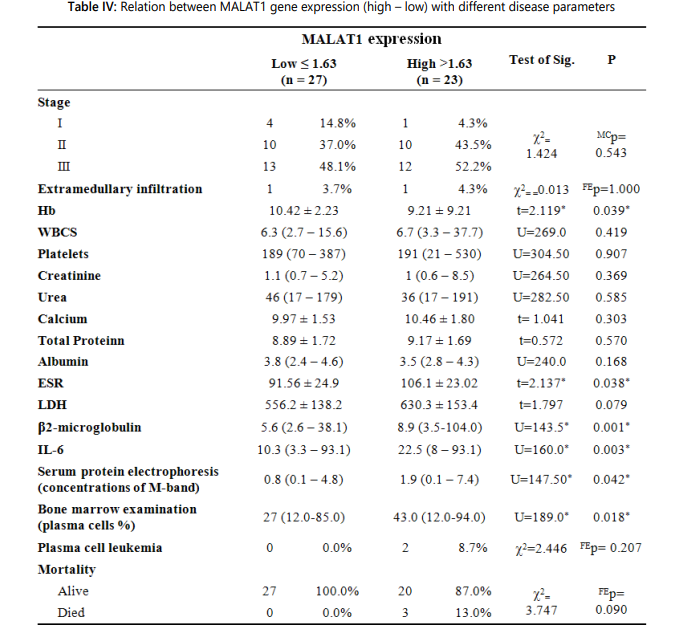
Furthermore, MM patients were divided into two groups with high and low expression by the median value of the MALAT1 gene expression level, which was 1.62. It was found that high MALAT1 expression levels were significantly associated with high ESR (first hour) levels, high PC percentages in BM, high serum β2- microglobulin levels, high serum IL-6 levels, high M-protein concentrations in SPEP, and low hemoglobin levels. Moreover, no statistically significant association between the high MALAT1 expression levels and ISS stage in MM patients was reported in MM patients as shown in table IV.
Discussion
The pathogenesis of multiple myeloma involves complex genetic and epigenetic events. Lately, lncRNAs are implicated in the pathogenesis of several diseases including cancer. Their level of expression is sufficiently potent to serve as a marker for diagnosis, classification, prognostic evaluation of malignancies and predictive assessment of adequacy of treatment [22]. Data concerning lncRNAs involvement in MM are expanding. Unlike other lncRNAs, MALAT1 is abundantly expressed and evolutionarily conserved in various mammalian species. Numerous studies have shown that MALAT1 plays a key role in the development and metastasis of various cancers [23,24].
In the present study, the expression of MALAT1 was significantly higher in the MM patients at diagnosis (T0) as well as after 2 cycles of bortezomib-based therapy (T1) compared with those levels reported after 4 cycles of bortezomib containing therapy (T2) in MM patients or control subjects. However, the differences were not statistically significant between the expression levels reported in T1 when compared to T2 patients. Worth noting is also that the expression levels of the MALAT1 gene in both cases that presented with extra-medullary infiltration in our study were high.
In the T0 group, the MALAT1 gene expression levels showed a statistically significant positive correlation only with M-protein concentration in SPEP but not to other disease parameters. The MALAT1 gene expression levels in the T1 group showed significant positive correlations with PC percentages in BM, M-protein concentration in SPEP, ESR, and serum levels of LDH, total protein, as well as IL-6. Lastly, its expression levels in the T2 group displayed no significant correlation to all disease parameters studied. Correlations between MALAT1 gene expression and extramedullary myeloma spread were not applicable as only two cases showed extramedullary spread.
Moreover, we divided the MM patients into 2 groups with high and low MALAT1 expression levels based on the median value of the MALAT1 gene expression level, which was 1.62. It was found that high MALAT1 expression levels were significantly associated with high ESR (first hour) levels, high PC percentages in BM, high serum β2-microglobulin levels, high serum IL-6 levels, high M-protein concentrations in SPEP, and low hemoglobin levels. Moreover, we found no statistically significant association between the high MALAT1 expression levels and ISS stage in MM patients.
These findings further suggest that MALAT1 may be deregulated and overexpressed in MMpatients and highlight its potential use as a novel prognostic biomarker for MM. Hand in hand with the findings of our study, Cho et al. measured the MALAT1 expression levels in 18 patients with relapsed or advanced MM, 45 newly diagnosed patients, and 61 patients after treatment and reported that NDMM patients expressed higher levels than the post-treatment patients or the healthy individuals. In their study, a strong correlation existed between disease status and MALAT1 expression levels as documented by its markedly decreased levels in the posttreatment patients to levels similar to those in the healthy subjects. In addition, in patients with progressive or relapsed disease, the MALAT1 expression levels were reported to be markedly increased in contrast to post-treatment patients. Cho et al., however, could not establish an association between MALAT1 expression levels and the percentage of PCs in BM, which is contrary to our observations. Moreover, they determined that magnitude of decrease in MALAT1 expression levels post-treatment was prognostic, as patients who progressed early showed significantly smaller decrease in expression levels, while patients with greater decrease in expression levels had significantly longer PFS [20]. However in the current study, the MALAT1 gene expression levels were only assessed once for every MM patient, which did not allow the assessment of the magnitude of the change (decrease) after myeloma-related therapy as demonstrated by Cho et al.
Similarly, another study by Ronchetti et al. demonstrated that 31 lncRNAs were deregulated in MM cells, including MALAT1. They also showed that the upregulation of MALAT1 in MM was associated with molecular pathways regulating cell cycle, p53-mediated DNA damage response, and mRNA maturation processes denoting that higher expression levels of MALAT1 were associated with MM disease progression [25].
In contrast to the findings of our study, Isin et al. reported significantly lower plasma MALAT1 levels in patients with multiple myeloma in contrast with healthy individuals. In this study, 58
MM patients were investigated, but it was unclear whether those patients were newly diagnosed or under treatment [26]. Different sample sources could be a possible cause to blame for this discrepancy. Since the disease pathogenesis is closely related to the BM microenvironment, we examined the MALAT1 expression levels in bone marrow samples and not in plasma samples. Furthermore, the higher MALAT1 expression levels encountered in our study may be attributable to the BM microenvironment that favors myeloma cell proliferation.
In accordance with our findings, Handa et al. demonstrated that the MALAT1 expression was significantly higher in BM PCs of MM (median level 6.745) compared to that in control (median level 1.983) cells. Of note, the median MALAT1 expression levels in both MM (1.62) and control (0.75) groups in our study were lower compared to those determined by Handa et al. These differences may be attributable to the small sample size, the differences in the study design and the selection of the population studied. Similar to our findings, they also demonstrated that the MALAT1 expression did not differ among different stages as classified by the ISS stage. Additionally, they investigated 114 MM patients for the association of MALAT1 with extramedullary myeloma (EMM). In contrast to bone marrow PCs, MALAT1 expression levels were found to be up to several thousand-fold upregulated in PCs from EMM obtained from the same patients in this pair wise comparison, thus proposing its potential role in extramedullary spread. This hypothesis was further supported by their correlation of higher MALAT1 expression with worse OS and PFS; in particular, MALAT1 expression showed an independent influence on PFS, suggesting a possible role in drug resistance [27]. In the present study, the correlation between MALAT1 gene expression and extramedullary myeloma spread was not applicable as only two cases showed extramedullary spread.
Identification of different roles of lncRNAs in MM will be essential step toward the comprehension of pathobiology and cancer-promoting pathways in MM. Furthermore; lncRNA may be utilized as excellent anticancer therapeutic tools, because of their cancer-specific expression that represent a major advantage over other therapeutic approaches [28]. Of note, MALAT1-silencing strategies led to inhibition of MM tumor growth, thus providing the preclinical rational for considering MALAT-1 a novel therapeutic target in MM [29-31].
In general, the discrepancies between the results of the current study and those reported by other authors may be attributable to the choice of population to be studied, the small patient sample, the gene-environment interactions, and the different study design. Therefore, further studies are required to confirm their prognostic potentials using larger groups of patients with long-term follow-up.
Conclusion
We have demonstrated that MALAT1 gene was overexpressed in newly diagnosed MM patients compared with post-treatment MM patients and control group. Additionally, there were statistically significant positive correlations between level of MALAT1 expression and plasma cell percentage in bone marrow, abnormal levels of IL-6, ESR and concentration of M-Band in serum protein electrophoresis. The current data supported the finding established by others that MALAT1 may be useful as a novel prognostic biomarker for MM. These may support the necessity to explore the efficiency of MALAT1 gene as a novel therapeutic target.
References
- Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, riskstratification and management. Am J Hematol 2020; 95: 548–567.
- Cowan AJ, Allen C, Barac A, et al. Global burden of multiple myeloma: A systematic analysis for the global burden of disease study 2016. JAMA Oncol 2018; 4(9): 1221–1227.
- AnnamariaGulla, Kenneth C Anderson. Multiple myeloma: The (r) evolution of current therapy and a glance into future. Haematologica 2020; 105(10): 2358-2366.
- Lu M, Hu Y, Wu Y, et al. Genome-wide discovery and characterization of long noncoding RNAs in patients with multiple myeloma. BMC Med Genomics 2019; 12(1): 135. doi:10.1186/s12920-019-0577-5
- Wang C, Wang L, Ding Y, et al. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci 2017; 18(12): 2659. doi: 10.3390/ijms18122659
- He Q, Liu Y, Sun W. Statistical analysis of non-coding RNA data. Cancer Lett. 2018; 417: 161-167. doi: 10.1016/j.canlet.2017.12.029
- Fachrul M, Utomo DH, Parikesit AA. lncRNA-based study of epigenetic regulations in diabetic peripheral neuropathy. In Silico Pharmacol. 2018; 6(1): 7. doi: 10.1007/s40203-018-0042-8
- Deng H, Wang JM, Li M, et al. Long non-coding RNAs: new biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol 2017; 39(6):1010428317706332. doi: 10.1177/1010428317706332
- Chen H, Xu Z, Liu X, et al. Increased expression of Lncrna RP11-397A154 in gastric cancer and its clinical significance. Ann Clin Lab Sci 2018; 48(6): 707–711.
- Nobili L, Lionetti M, Neri A. Long non-coding RNAs in normal and malignant hematopoiesis. Oncotarget 2016; 7:50666-50681. doi: 10.18632/oncotarget.9308
- Bonetti A, Carninci P. From bench to bedside: the long journey of long non-coding RNAs. Curr Opin Syst Biol. 2017; 3: 119–124. doi: 10.1016/j.coisb.2017.04.016
- Ronchetti D, Agnelli L, Pietrelli A, et al. A compendium of long noncoding RNAs transcriptional fingerprint in multiple myeloma. Sci Rep. 2018; 8: 6557. doi: 10.1038/s41598-018-24701-8
- Wong NK, Huang CL, Islam R, et al. Long non-coding RNAs in hematological malignancies: translating basic techniques into diagnostic and therapeutic strategies. J Hematol Oncol. 2018; 11: 131. doi: 10.1186/s13045-018-0673-6
- Zhao M, Wang S, Li Q, et al. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol Lett. 2018; 16(1): 19-26.
- Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 2017; 490(2): 406-414. doi: 10.1016/j.bbrc.2017.06.055
- Ragusa M, Barbagallo C, Statello L, et al. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015; 21: 11709-11739. doi: 10.3748/wjg.v21.i41.11709
- Rajkumar SV. Updated Diagnostic Criteria and Staging System for Multiple Myeloma. Am Soc Clin Oncol Educ Book 2016; 35: e418-23. doi: 10.1200/EDBK_159009
- Kumar S, Paiva B, Anderson KC, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016; 17(8): e328-e346. doi: 10.1016/S1470-2045(16)30206-6
- Prieto B, Miguel D, Costa M, et al. New quantitative electrochemiluminescence method (ECLIA) for interleukin-6 (IL-6) measurement. Clin Chem Lab Med. 2010; 48(6): 835-8. doi: 10.1515/CCLM.2010.153
- Cho SF, Chang YC, Chang CS, et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014; 14:809. doi: 10.1186/1471-2407-14-809
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods. 2001; 25 (4): 402–8. doi: 10.1006/meth.2001.1262
- Arun G, Aggarwal D, Spector DL. MALAT1 Long Non-Coding RNA: Functional Implications. Noncoding RNA. 2020; 6(2): 22. doi: 10.3390/ncrna6020022
- Butova R, Vychytilova-Faltejskova P, Souckova A, Sevcikova S, Hajek R. Long Non-Coding RNAs in Multiple Myeloma. Noncoding RNA. 2019; 5(1): 13. doi: 10.3390/ncrna5010013
- Cui YS, Song YP, Fang BJ. The role of long non-coding RNAs in multiple myeloma. Eur J Haematol. 2019; 103 (1): 3–9. doi: 10.1111/ejh.13237
- Ronchetti D, Agnelli L, Taiana E, et al. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget. 2016; 7 (12): 14814–14830. doi: 10.18632/oncotarget.7442
- Isin M, Ozgur E, Cetin G, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014; 431: 255-259. doi: 10.1016/j.cca.2014.02.010
- Handa H, Kuroda Y, Kimura K, et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br J Haematol. 2017; 179(3): 449–460. doi: 10.1111/bjh.14882
- Nobili L, Ronchetti D, Agnelli L, et al. Long non-coding RNAs in multiple myeloma. Genes. 2018; 9(2): 69. doi: 10.3390/genes9020069
- Amodio N, Stamato MA, Juli G, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018; 32(9): 1948–1957. doi: 10.1038/s41375-018-0067-3
- Amodio N, Raimondi L, Juli G, et al. MALAT1: A druggable long noncoding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018; 11(1): 63. doi:10.1186/s13045-018-0606-4
- Liu H, Wang H, Wu B, et al. Down-regulation of long non-coding RNA MALAT1 by RNA interference inhibits proliferation and induces apoptosis in multiple myeloma. Clin Exp Pharmacol Physiol. 2017; 44(10): 1032-1041. doi: 10.1111/1440-1681.12804


