University of Calcutta, India
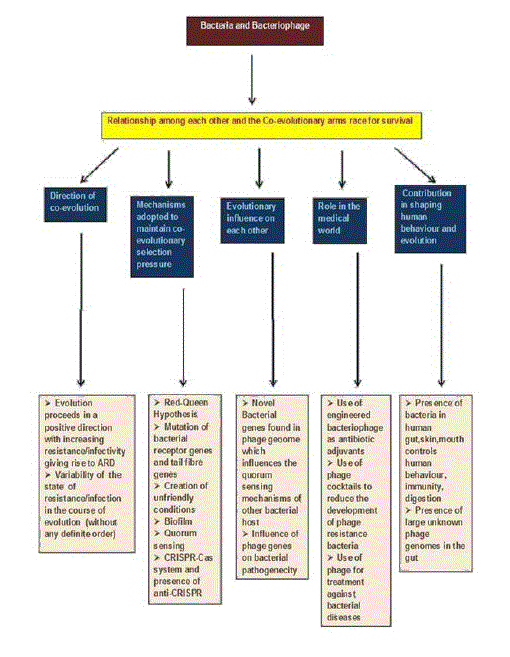
Background: The first organisms to appear on the earth were unicellular prokaryotes from where the present day bacteria evolved. The origin of virus is not yet known. Viruses may have evolved from bacteria or plasmids. In order to survive bacteria has to prove their mettle against a type of viruses called bacteriophages which infects the bacterial cell and kills it. In course of time bacteria evolved different mechanisms to evade the infection by bacteriophages and phages also developed ways to infect these resistant bacteria. This constant co-evolutionary arms race for existence led to the diversification of bacterial and bacteriophage species with advanced adaptive features. This review deals about the antagonistic co-evolutionary relationship of phage and bacteria, impact of it on each other and on the life of humans.
Methods: The review paper is written after reading a list of research papers from the NCBI (PubMed) database.
Results: The co-evolutionary arms race generally follows a directional route of evolution with increase in resistance/infectivity. But there are also fluctuations in the state of infection/resistance in course of evolution where there is no fixed order of increase or decrease of the infectivity and resistance of the parasite and host respectively. Bacteria employ different methods for protection against phage attack and the phages also adopt different mechanisms to overcome bacterial resistance.
Novel bacterial genes are found in phage genome which influence quorum sensing of other bacteria. It was also seen presence of phage genes controls pathogenicity of bacteria. These findings showed the evolutionary impact of the organisms on each other.
Phages can be used as therapeutic agents against bacterial infection. Apart from this application, a large amount of unknown phage genomes were discovered in human gut whose probable role in shaping human behaviour are yet to be revealed.
Conclusion: The co-evolutionary struggle for existence paved the way for diverse group of organisms of phage and bacteria with increased adaptive characters. A part from the antagonistic relationship both the organisms have influence on each other which is of evolutionary significance.
Biography:
Aparajito Sen from Kolkata, India and he applying for a paper presentation in the symposium in Valencia, Spain on Microbiology. He completed his Masterʼs degree in Genetics in 2017 from University of Calcutta, Kolkata, India and wish to pursue PhD in future. He completed his Bachelorʼs degree in Botany from Scottish Church College affiliated to University of Calcutta and he completed his school level secondary and higher secondary examination from St Lawrence High School, Kolkata India. His medium of education was English throughout. He is very fluent in English from both speaking and writing aspects. He want to present a review paper on co-evolutionary arms race between bacteria and phages which he wrote in the final semester of his Masters as a dissertation project. He is currently working in University of Calcutta as a research trainee in Dr Sreya Chattopadhyayʼs lab (Cancer biology lab) in Department of Physiology, University of Calcutta, India.
Padova University School of Food Science, Italy
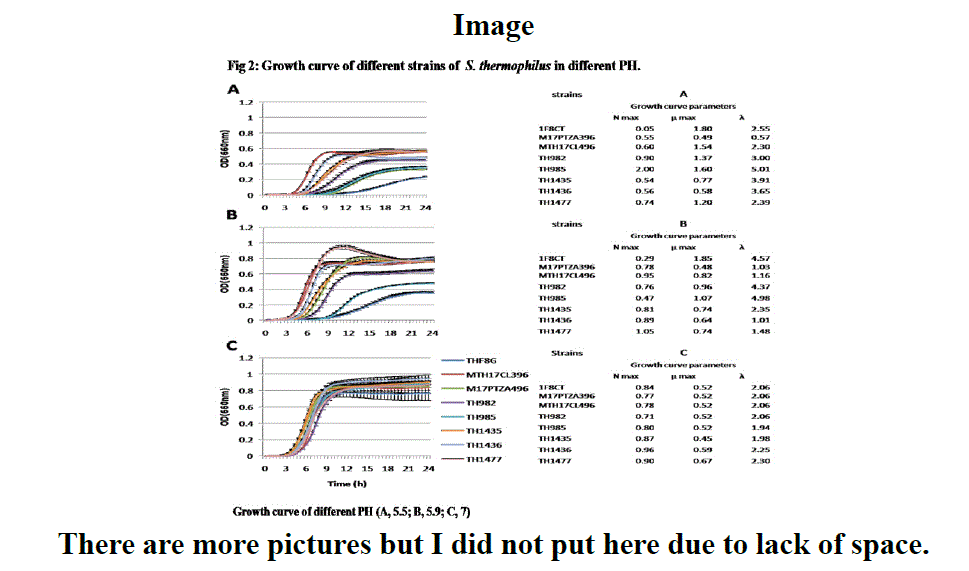
The aim of this study was to discover more about S. thermophilus and its abilities by comparing it with a very less-known bacterium like S. Macedonicus. As a member of the LAB, Streptococcus thermophilus is identified as a thermophilus group and use as a commercial starter in industry. On the other hand, although Streptococcus Macedonicus was first described more than 16 years ago but still the exact role of this bacteria in the industry is not distinct. In this study, eight new isolated strains of S. thermophilus were applied to be investigated from the technological point of view. Analysis such as growth in different pH temperature and sugars, Antimicrobial susceptibility test and Inhibitory activity test were performed and the results were evaluated by using General linear model (GLM) in SAS 9.4. In all the comparison like growing in different PH, temperature and sugars between these two species, S. Macedonicus indicated the better growth rate which shows more functional bacteria. Image
Recent Publications (minimum 3)
1. Publish article on advance study in biology journal (ISI) as a corresponding author. (Investigation of resistance to the antibiotic penicillin, cefteriaxone, ciprofloxacin and cefixime in neisseria gonorrhoeae sampled frompatients infected with the gonorrhea in Poona city, India)
2. Publish article on advance study in biology journal (ISI) as a corresponding author (Frequency of the Pathogenic Campylobacter in the Swab-Rectal Samples of the Seagulls(Larus Canus) Using the PCR Technique).
3. Publish article on research journal of fisheries and hydrobiology as a corresponding author(ISI).
4. Publish article on international journal of biology, pharmacy and allied science (ISI) (Investigation of changes in levels of Serum Elements, Lipid Profile and Advanced Glycation End Product in patients with type 2 diabetes)
Biography:
Armin Tarrah student at Padova University in Food Microbiology. We are involved in fermentation and lactic acid bacteria nowadays and also we work on probiotics area. Tarrah bachelor and master degree in cellular and molecular biology-Microbiology as well. Tarrah was also University teacher at Chaloos azad university. Tarrah tought microbiology, bacteriolog at medical department at this university. Tarrah cooperating as a reviewer with international journal of food microbiology in Netherland as well.
Biotechnology Research Institute, China
Despite extensive vaccination, H9 Avian influenza outbreaks has caused great economic losses to poultry industry resulting in decrease egg production, high morbidity and mortality. The ability to cross species barrier makes it a potent threat. Continuous mutations in the HA gene transforms AIV subtype H9N2 into more pathogenic virus that may have pandemic potential and can cross species barrier. Thus, it is essential to continuously monitor antigenic variants of H9 virus. HA gene plays vital role in viral attachment, release of genetic material and pathogenicity. In present study, a sum of four H9 virus samples were isolated, serological and molecular confirmation was done.500samples were collected and properly labelled. They were then processed for egg inoculation in embryonated eggs. Virus was grown in embryonated eggs and harvested fluid is then preceded for confirmatory testing. Haemagllutination and Haemagllutination Inhibition testing was done. RNA was extracted by Kit method and cDNA was synthesized. Reverse Transcriptase (RT-PCR) was performed using specific primer sets and then the amplicon were run on agarose gel. PCR product was sent for sequencing and Phylogenetic tree was constructed. The present study enabled us to characterize and construct Phylogenetic tree of HA gene of currently prevailing H9N2 Avian Influenza isolates in Pakistan belongs to G1-like sub lineage. Amino acid analysis revealed substitution of polybasic amino acid residues that my transform H9N2 into more pathogenic.
1ARC-Onderstepoort Veterinary Research, South Africa
2University of the Western Cape, South Africa
3ARC-Plant Protection Research, Pretoria
Aspergillus flavus is the main producer of carcinogenic aflatoxins (AFB1 and AFB2) in agricultural commodities such as maize. The purpose was to investigate four strains of A. flavus for aflatoxins production. The strains: 3909, 3911, 3951 and 3955 isolated in Mpumalanga were morphologically identified at ARC-Plant Protection Research Institute and further characterised by Polymerase Chain Reaction (PCR) and rDNA region of ITS-5, 8-ITS2. They were also analysed for the presence of genes encoding AFB1, targeting both regulatory (aflR, aflS) and structural genes (aflD, aflM, aflO, aflP and aflQ). A reverse high performance liquid chromatographic (HPLC) instrument was used for aflatoxin analysis. All the four strains amplified 600bp of ITS-5. 8-ITS2 rDNA region. Similarly, all genes for aflotoxin B1 were detected in four strains with expected band sizes. Aflatoxin production was present in strain 3911 and 3955 for AFB1 and AFB2 and in strain 3951 only AFB1 while strain 3909 revealed negative aflatoxin (AFB1 and AFB2) production. The results may contribute to development of reliable molecular techniques for detection of aflatoxigenicity as well as illustrating the complexity of local fungal communities associated with maize.
Biography:
Athini Ntloko is currently a final year PhD student under Professional Development Programme (PDP) at Agricultural Research Council (ARC) and registered with University of the Western Cape in 2016 and conducting a research project in the study entitled: Evaluation of the capacity of hydrogen sulphide to impact infection and aflatoxin contamination of maize by Aspergillus flavus. In 2017, she has been awarded a third prize for poster presentation from an annually ARC PDP conference. Ms Ntloko earned her Bsc degree (Microbiology and Biochemistry) in 2013, honours (Microbiology) in 2014 and Masters in Microbiology (2016) with University of Fort Hare.
1Department of Agro Economics, Mizan-Tepi University, Ethiopia
2Department of Natural Resource Economics, Mizan-Tepi University, Ethiopia
3Department of Horticulture, Mizan-Tepi University, Ethiopia
Ethiopia has possibly the highest potential for potato production than any country in Africa. Postharvest loss (20–25%) is one of the major problems in the potato production. Therefore, this study was conducted with the objective of assessing postharvest losses along potato value chain actors and identifying its determinants in the study area. The descriptive result indicated that the quantity of postharvest losses at producer, local trader, and whole saler and retailer level was 21.724, 1.838, 3.406 and 4.07 kg/qt, respectively. The average gross margin with loss of producerʼs local traders, wholesalers and retailers was 6464.70, 282,169.89, 219,644.61 and 345,826.36 Birr, respectively, which is less than the average gross margin without loss (10,146.12, 284,015.83, 221,274.69 and 352,986.62 Birr, respectively). Distance to the nearest market, area allocated for potato and total output determine postharvest loss positively, and sex, experience, family size of working age, selling price and access to credit determine postharvest loss negatively. In the study area, lack of storage facilities for potato was raised by farmers and other actors as a priority problem. Intervention of government from input supply until the end consumers is paramount and preparing storage mechanism is a must.
Keywords: Gross margin, multiple linear regressions, Postharvest loss, Potato
Biography:
Benyam Tadesse born on 02-12-87. Benyam Tadesse graduated from Hawassa University from the department of Agricultural resource economics and management. Then Benyam Tadesse joined Jimma University to attend MSc in Agribusiness and value chain management and successfully accomplished. Know Benyam Tadesse Lecturer, Researcher and director at Mizan Tepi University. Benyam Tadesse published 5 manuscripts & happy to participate on the occasion.
Birla Institute of Technology and Science, India
Seven putative Na+/H+ antiporters have been reported to be present in A. platensis NIES-39 which thrives well even at pH 11. This raises a question whether all seven antiporters are equally involved in alkaline pH homeostasis in this cyanobacterial strain. To this end, we studied the transcriptional profile of these seven putative Na+/H+ antiporters. We found a significant difference in the m-RNA levels of the seven antiporters at pH 7, 9 and at 11. Also, a temporal pattern of the expression profile was observed. In silico conserved domain analysis of these seven putative antiporters indicated the presence of nine different kinds of domains in all, out of these nine domains, six domains function as monovalent cation-proton antiporters and two of the domains function as the universal stress protein (Usp) category which are UspA and Usp Like. The protein sequence of these putative antiporters have been compared and correlated with the solved crystal structure available from the CPA Super family (CPA1 & CPA2). The In-silico analysis and the real time PCR analysis, put together, suggest an active participation of these seven putative Na+/H+ antiporters in alkaline pH homeostasis of this cyanobacterial strain.
Biography:
Bhagavatula Vani is presently working as Assistant Professor in Department of Biological sciences, BITS, Pilani, India. She has carried out her Doctoral in the field of abiotic stress and photosynthesis in Rice plants under the supervision of Late. Prof. Prasanna Mohanty, JNU, India who is an exponent of photosynthesis from India. She has published some of her works in journals like JPP, Photosynthetica, Plant and Soil and BiologiaPlantarum. She has also worked on nitrogen metabolism and abiotic stress in the wonder microbe, Arthrospira platensis (Spirulina platensis) and published some of the results.
Chukwuemeka Odumegwu Ojukwu University, Nigeria
Biodegradation of crude-oil polluted soil by was carried out to determine the ability of bacteria to utilize and detoxify crude oilpolluted soils. Samples were collected from crude oil-polluted environments of Niger Delta ecological zone. Physico-chemical, microbiological, adaptability tests, biochemical and molecular characterization of the isolates were assayed. Composite samples were collected from a depth of 20cm, soil pH, moisture content, water holding capacity and total organic carbon was carried out. Serial dilution using spread plate method was used to determine the total heterotrophic bacteria and hydrocarbon utilizing bacteria. Residual total petroleum hydrocarbon was analyzed using gas chromatographic flame ionization detector method. Identity of bacterial isolates was confirmed using polymerase chain reaction techniques. Results revealed that pH ranged between 3.65–6.1, total organic carbon content ranged from 3.4–7.1 %, total viable counts ranged from 7.1 x 106–9x1010, the bacterial load was high for all sample. Environmental indigenous bacteria were able to mineralize the crude oil in the soil. Adaptability test revealed values ranging from 0.11–0.54, all the isolates adapted well to crude oil exposure. Biochemical test results revealed Bacillus spp., Pseudomonas spp., Serratia spp., Micrococcus spp., Bacillus spp., Proteus spp., Arthrobacter spp. and Shigella spp. PCR results gave the identity of the organisms as Lysinibacillus spp. M2c, Serratia marcescens Mb4, Bacillus aerius TPM-23, Proteus mirabilis LS-3, a new isolate was encountered. TPH concentrations were in the range of 123.67 mg/l–753.32 mg/l. This study has established the ability of the isolates; Serratia marcescens Mb4 and Lysinibacillus spp. M2c to utilize and detoxify crude oil-contaminated soils.
Biography:
Umeaku Chinyelu has completed her Ph.D at the age of 52 from Federal University of Technology, Owerri, Nigeria. She has a diploma of the National Institute of Science Laboratory Technology. She is a member of the Organization for Women in Science for the Developing World (OWSD), a member of Science Teachersʼ Association of Nigeria (STAN), a member of Nigerian Society for Microbiology (NSM), a member of American Society for Microbiology (ASM). She is an author and has published many journal articles and attended several conferences both locally and internationally.
Nile University of Nigeria, Nigeria
Developments in molecular biology have affected whole fields of biology positively in recent years. These developments have also showed their effects on fungal systematic and provided fast and reliable identifications. So, molecular techniques have started to be used commonly with conventional techniques in fungal systematic. Some of these are used in common recently, and those based on PCR techniques and especially DNA sequences studies.
Amatoxins are potent inhibitors of RNA polymerase II, indirectly halting protein synthesis. It causes the death of the cell. Amatoxins are the main poison of the green death cap (A. phalloides) and among the most dangerous natural toxins causing hepatic failure. Fatal mushroom poisonings are predominantly caused by members of the Amanita family, including the genera A. phalloides, A. virosa, and A. verna.
Identification of amatoxin gene that belongs to Amanita genus is responsible for >90% of mushroom poisoning by using PCR techniques and DNA sequences analyses constitutes the aim of this study.
In this study we aim to identify and compare genes encoding Amotoxin by designing primers belong to the amotoxin. While conducting this study, the A. phalloides, A. virosa and A. verna species will be used and samples belong to Turkey and U.S. This study ensures to find out the differences of gene encoding amatoxin between the Amanita genus. The results of this study can help the cancer treatments by making some modifications to these toxin genes by recombinant DNA technologies, only cancer or tumor cells can be destroyed with modified proteins instead of healthy cells.
Biography:
Hatice Tetik born in Erzurum, Turkey in 1981. Hatice Tetik obtained bachelor degree in 2005 with first degree award in Biology at Canakkale 18 March University in Turkey. After graduation, Hatice worked as a teacher at different private educational institutions in Turkey for 3 years and then started Master of Science in Biology because of interest in academic studies. Currently Hatice has completed coursework and studying on thesis at Nile University of Nigeria.
University of Nottingham, UK
The depletion in fossil fuel reserves has instigated the search for renewable sources such as biofuels. Biofuel production provides a sustainable alternative to fossil fuels. However, the progression of biofuel industry has been largely affected by several uncertainties including the sustainability of production processes. Finding an economically viable process and the right substrates have been a source of constant debate. Biodiesel production is one such area which is gaining momentum in the last few decades. We are currently investigating cassava wastewater
As a substrate and also as a source of oleaginous bacteria. The production of tri-acylglycerol from mycolic acid containing actinomycetes predominantly Rhodococcus, has been identified as a potential source. These bacteria which are largely ubiquitous provide a significant amount of triglyceride when cultivated under low cost waste. The growth of oleaginous bacteria for the accumulation of triglycerides on low cost and abundant, cassava waste still remains unexplored, especially in Nigeria which is the largest producer of cassava in the world. We started our initial investigations on the cassava waste water sample following culture dependent and independent approach. We prognosticate that the identification of microflora isolated from different stages in the sample will provide a basis for understanding the nature of the substrate and the potential for the synthesis of biodiesel from cassava waste water.
1,2Jimma University, Ethiopia
3Vrije Universiteit Brussel, Belgium
Background: Giardia duodenalis imposes significant public health crisis on developing countries, however its genetic diversity and infection risk factors are less studied in these areas. This study was conducted to determine the molecular epidemiology of G. duodenalisin Southern Ethiopia.
Methods: From March to June 2014, stool samples were collected from 590 randomly selected individuals. Socio-demographic data were gathered using a pre-tested structured questionnaire. The genotyping was done using TPI gene-based nested PCR and DNA sequencing. The isolates genetic identity and relatedness were determined using BLAST and phylogenetic analyses. Risk factors were analysed using binary and multinomial logistic regressions.
Results: The results showed that 18.1% (92/509) of the subjects were infected by G. duodenalis, 35.9% (33/92) was assemblage A, 21.7% (20/92) assemblage B and 42.4% (39/92) mixed infections of A and B. Most assemblage A isolates (94%, 31/33) were 100% identical to sequences in GenBank, the majority belonged to sub-assemblage AII. However, the high frequency of double peaks in chromatogram made assemblage B sub-typing more problematic, only 20% (4/20) of the isolates matched 100% with the sequences. Age (P=0.032) and drinking water sources (P=0.003) showed significant association with G. duodenalis infection.
Conclusion: G. duodenalis infection is endemic in Southern Ethiopia; assemblage A was more frequent than assemblage B. The rate of infection was higher in children and in municipal/tap and open spring water consumers than the other groups. The present study confirms the need for further studies to be conducted focusing on sub-types of assemblage B and the origin of heterogeneity.
Biography:
Mengistu Damitie received his Bachelor Degree in Biology from Bahir Dar University in 2006 and Master of Science in Biology (Biomedical Sciences) from Addis Ababa University in 2010. He has been an instructor of Medical Parasitology, Immunology and Vector Biology from 2010/11-2013 in School of Medicine, College of Medicine and Health Sciences, Arba Minch University, Ethiopia. During this period, he was also served as head of School of Medicine and involved in different positions and professional memberships. Since 2014, he is doing PhD (Doctor of Science) in Vrije University Brussel, Belgium and Environmental Health in Jimma University, Ethiopia.
1Mutah University, Jordan
2Royal Medical Services, King Hussein Medical Center
3Aarhus University, Denmark
4Max Planck Institute of Infection Biology, Germany
Background & Aims: Helicobacter pylori (H. pylori) have been associated with gastritis, gastric ulcer, mucosa-associated lymphoid tissue lymphoma and gastric cancer. The prevalence of H. pylori virulence genes has been studied in different populations and from different sources of samples but their prevalence has not been studied in dental plaque in Jordanian people; therefore, the aim of this study was to determine the genotypes of H. pylori isolated from dental plaque samples.
Methods: Dental plaque samples were collected from 60 Jordanian volunteers. The genotypes of H. pylori virulence genes including the cytotoxin-associated gene (cagA) and the vacuolating toxin (vacA) were determined using polymerase chain reaction (PCR).
Results and Conclusions: The cagA gene was detected in 14 (23.3%) samples, while vacA was detected in all volunteers enrolled in this study (100%). The most prevalent vacA alleles were m2 and s1 in 54 (90%) and 55 (91.7%) of volunteers, respectively. Compared to the other combinations including the most virulen vacA genotype s1/m1 which was detected in 11 (18.2%) of volunteers, the most prevalent vacA allelic combinations were s1/m2 and s2/m2 in 56 (93.3%) and 27 (45%) of volunteers, respectively. These results indicate a significant carriage of virulent H. pylori strains among Jordanian people in their dental plaques, which increases the possible transmission of these strains among them. In addition, the studying of the genotypic pattern of H. pylori virulence genes in the dental plaque could represent an essential tool for infection prevention and predicting the severity and prognosis of H. pylori gastric infection.
1Damanhour University, Egypt
2Alexandria University, Egypt
A wide diversity of organisms exists in soil. Well-adapted groups can be found in extreme environments. Hypersaline ecosystems show a considerably rich diversity of microbes and are biologically very productive. These environments are rich source for novel microbes to be discovered. A newly isolated Salinicoccus sp. HMS from a soil sample of Wadi EL Natrun was characterized. A mucoid, glistening, reddish-orange pigmented colonies on Horikoshi-I agar plates was identified as Salinicoccus sp. HMS by morphological, physiological and biochemical characterization and 16S rDNA sequencing. S.sp. HMS grew in presence of 0–25% (w/v) of NaCl and at pH 6–11, with optimum growth at 11.7% (w/v) NaCl and pH 9. S.sp. HMS participates in halite formation in Horikoshi-I both supplemented with 2M, 11.7% (w/v) NaCl at pH 8. The bacterium was shown to produce a wide variety of extracellular hydrolytic enzymes including, inulinase, pectinase, amylase, lipase, mannanase, protease, cellulase, and xylanase. The strain was found to be capable to degrade phenol and chlorpyrifos. Moreover, the isolate due to its biopolymer, pigment and enzymes production under extreme condition displays potential biotechnological and bioremediation applications.
Keywords: Salinicoccus sp. HMS; Halophilic; Extracellular Enzymes; Biopolymer production
Biography:
Mona E. M. Mabrouk Professor of Microbiology. She is working at Botany and Microbiology Department, Faculty of Science, Damanhour University, Damanhour, Egypt. Her research of interests is Microbial Biotechnology.
1Technion-Israel Institute of Technology, Israel
2University Grenoble Alpes-National Center for Scientific Research (CNRS)-Commissariat for Atomic Energy and Alternative Energies (CEA), France
Amyloids are structured protein fibrils associated with human fatal neurodegenerative diseases. Recently, microbial amyloids were found to be ubiquitous across kingdoms of life and to perform many physiological functions, including as key virulence determinants in microbes. These activities include stabilizing microbial biofilms, mediating host-pathogen interactions and competing with other bacteria. In this study we shed light on the functional and structural roles of amyloid-forming peptides called Phenol Soluble Modulins (PSMs) secreted by the pathogenic Staphylococcus aureus. As a means of pathogenesis, PSMs cause lysis of human cells including leukocytes and erythrocytes, stimulate inflammatory responses and contribute to biofilm development particularly in virulent S. aureus strains such as the community-associated methicillin-resistant S. aureus (CA-MRSA). We found that PSMα1 and PSMα4, involved in biofilm structuring, form cross-β amyloid fibrils that were linked with eukaryotic amyloid pathologies, shown here for the first time at atomic resolution in bacteria. These fibrils confer high stability to the biofilm. In contrast, the cytotoxic activity of PSMα3 against human cells stems from the formation of cross-α fibrils (Tayeb-Fligelman et. al., Science 2017) that are at variance with the cross-β fibrils. Interestingly, a truncated PSMα3, which forms reversible fibrils and has antibacterial activity, reveals two polymorphic and a typical β-rich fibril architectures, both radically different from both the cross-α fibrils formed by full-length PSMα3 and from the cross-β fibrils formed by PSMα1 and PSMα4 (Salinas et. al., Nature Communications, in press). Our results point to structural plasticity being at the basis of functional diversity exhibited by S. aureus PSMαs.
Biography:
Nir Salinas is a doctoral student in the Structure Function Relationships in Microbial Functional Amyloidsʼ lab, faculty of biology at the Technion-Israel Institute of Technology, under the supervision of Assist. Prof. Meytal Landau. Nir Salinas Ph.D. studies determine the fascinating and unexplored structures of bacterial functional amyloids, correlate the structures to their functions as key virulence determinants, and device means for their regulation. The ultimate goal of Nir Salinas research is to develop novel antivirulence and antibacterial drugs with a new mechanism of action based on Amyloidogenic peptides.
University of Trento, Italy
Water springs provide important ecosystem services including drinking water supply, recreation and balneotherapy, but their microbial communities remain largely unknown. In this study we characterized the spring water microbiome of ComanoTerme (Italy) at four sampling points of the thermal spa, including natural (spring and well) and human built (storage tank, bathtubs) environments. We integrated large-scale culturing and metagenomics approaches, with the aim of comprehensively determining the spring water taxonomic composition and functional potential.
The groundwater feeding the spring hosted the most atypical microbiome, including many taxa known to be recalcitrant to cultivation. The core microbiome included the orders Sphingomonadales, Rhizobiales, Caulobacterales and the families Bradyrhizobiaceae and Moraxellaceae. A comparative genomic analysis of 72 isolates and 30 metagenome-assembled genomes (MAGs) revealed that most isolates and MAGs belonged to new species or higher taxonomic ranks widely distributed in the microbial tree of life. Average Nucleotide Identity (ANI) values calculated for each isolated or assembled genome showed that 10 genomes belonged to known bacterial species (>95% ANI), 36 genomes (including 1 MAG) had ANI values ranging 85-92.5% and could be assigned as undescribed species belonging to known genera, while the remaining 55 genomes had lower ANI values (<85%). A number of functional features were significantly over- or under- represented in genomes derived from the four sampling sites. Functional specialization was found between sites, with for example methanogenesis being unique to groundwater whereas methanotrophy was found in all samples.
Current knowledge on aquatic microbiomes is essentially based on surface or human associated environments. We started uncovering the spring water microbiome, highlighting an unexpected diversity that should be further investigated. This study confirms that groundwater environments host highly adapted, stable microbial communities composed of many unknown taxa, even among the culturable fraction.
Central Marine Fisheries Research Institute, India
Tetrodotoxin (TTX) is a non-protein neurotoxin that was mainly isolated from many species of puffer fishes. TTX isolated from the diverse groups of organisms were totally unrelated to each other phylogenically. Four cases of food poisoning in Thailand after the consumption of horseshoe crab eggs, Carcinoscorpius rotundicauda, were reported, which was later confirmed to contain TTX. The TTX occurrence in gastropod was first reported in Japan, due to the consumption of a Trumpet shell, Charoniasauliae caused the paralytic food poisoning (PFP) in 1979, and the responsible toxin was identified as TTX. TTX was isolated from the Japanese Ivory shell, Babylonica japonica. Later, Noguchi (1982) put forward that the exogenous origin of TTX among animals through the food chain, the trumpet shell toxification through the consumption of the toxic star fish, Astropecten polyacanthus, so it was not support proposal of endogenous origin of TTX. Also TTX was being isolated from variety of other animals including xanthid crabs, Atergatiusfloridus, Atergatopsisgermanini and Zosimusaeneus, the marine flatworms, Planoceramultitentaculata, the marine arrow worms (Zooplankton) such as Eukrohina hamate, Parasagittaelegans and Flaccisagitascrippsae, which also play an important role in the food web to intoxicate the higher. Most of the TTX-bearing animals mentioned above have one common feature that they all belong to marine environment except the salamander and frogs and they are inter-related within a marine food-web system, in which TTX is accumulated and concentrated from lower to higher organisms through the food chain. Based on the above suggestion, that bacteria may be the primary source of TTX. In 1980, the first TTX-producing microorganisms were isolated and identified from TTX-bearing organisms by the Japanese scientists. Noguchi et al. (1983), succeeded in isolating four bacterial strains (Vibrio sp.) from the intestine of Xanthid crab, Atergatiusfloridus, and Yasumoto et al. also isolated a bacterial species Pseudomonas sp., from the marine red calcareous algae, Jania sp.. Therefore, many TTX-producing bacteria are isolated from different TTX bearing organisms. There were totally seven dominant pure bacterial strains that are isolated from different organs of puffer fishes and only five of them designated (Table-5.1), as although biochemical tests revealed the general characteristics of the bacterial strains. All the isolates showed 99% similarity with the available database sequences and then the phylogenetic trees, were constructed with the help of sequencing data. The five purified strains were Isolates 1 with Bacillus humi; Isolates 2 with Kocuriaplaustris; Isolates 3 with Pseudomonas mendocina; Isolates 4 with Pseudomonas stutzeri and Isolates 5 with Staphylococcus haemolyticus. In the LC50 data, the effect of different concentrations of five isolates viz 1000ppm, 500ppm, 250ppm, 125ppm, 62.5ppm and 32.25ppm were evaluated against Aretima bioassay. The highest Artemiamortality was found in 1000ppm concentration and the lowest mortality was recorded at 32.25ppm. The increase in concentration of TTX-producing bacteria sample was found to be increase the total mortality of Artemia. EPA Probit Analysis Program used for calculating LC50 value. The result of LC50 value, Isolates 1 was 209.159µg/ml, Isolates 2 was 317.007µg/ml, Isolates 3 was 260.365µg/ml, Isolates 4 was 149.793µg/ml and Isolates 5 was 150.024µg/ml. The purified strains, using the phenotypic characteristic as well as some additional physiological and biochemical test for genus level identification, were compared with results obtained from the 16s RNA sequencing. For the nucleotide sequence accession numbers, the partial 16s derived in this study have been deposited in Gen Bank under the accession numbers KP324953, KP324952, KP324955, KP324954 and KP324951.
1CSIR Institute of Microbial Technology, India
2National Institutes of Pharmaceutical Education and Research, India
Many Eukaryotic and Prokaryotic pathogens build infection in mammals including humans by netting the Proteolytic enzyme Plasminogen (Plg) onto their surface to digest host extracellular matrix (ECM). Lots of multifunctional glycolytic enzymes like glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Enolase have been reported in literature which interacts with host Plasminogen. In a self-protective response, the host steeds an inflammatory reaction, leading to infiltration of leukocytes to sites of inflammation, Macrophage infiltration from blood in proteolytically dependent manner utilizing Plasminogen and pathogen receptors ultimately promoting metastasis of pathogens to other parts of host. So there is constant struggle between host and pathogen to win the race against each otherʼs survival. Utilizing a combination of biochemical, cellular, knockdown, and In-vivo approaches, we would be demonstrating the partner/receptors present on pathogens for human Plasminogen to aid in their pathogenicity and simultaneously combat by host Plasminogen mediated host defense. As it is widely reported that Glycolytic enzymes have myriad of functions even in hosts as well as pathogens, we would try to explain the mechanism behind the host Plasminogen mediated Pathogenic aggression into host. Pathogen used in this study is Staphyloccocus aureus, Microcoocus luteus, Candida glabrata, Candida lusitaniae. We would be utilizing Solid Phase Binding ELISA, Pull Down Assay, HPG overlay assay, FACS, Confocal Microscopy to confirm interaction between Lab purified Human-Plasminogen; SDS-PAGE, LC-MS, MS/MS, to characterize the putative binding partner and finally cloning, Expression, Solid phase Binding assay, Surface Plasmon Resonance, to validate interaction between purified HPG and purified pathogenic receptor protein and finally Isothermal Colorimetry, Analytical Ultra Centrifugation for Stoichiometry judgment and lastly Side directed mutagenesis (Lysine mutants) as Plasminogen is known to interact with lysine residues of receptor molecules, cross linking trials, SAXS would be helping in throwing light on the interacting amino acid residues between two proteins. This whole study would be extremely helpful in alienating drug candidates against the pathogenic receptor protein to inhibit the pathogenesis via plasminogen interaction.
Biography:
Rahul Dilawari is pursuing his PhD dissertation under the supervision of Dr. Manoj Raje in a Cell Biology at the Institute of Microbial Technology (CSIR), Chandigarh. Our groupʼs research focus is primarily in the area of protein trafficking and protein-protein interaction. He possesses research experience of Molecular Biology, Microbiological, Bioinformatics tools, and recombinant DNA techniques for 4 years and 8 months, post Masterʼs degree before joining PhD in 2016. He has trained in PCR, Gene Cloning, heterologous protein expression, and Purification and Characterization techniques. This Conference exposure and interaction with some of the leading scientists of the field in India and abroad will assist & guide to develop and cement his skills in this area which would be useful for his long term goals of a future career in research.
Central University of Rajasthan, India
Visceral leishmaniasis (VL) is a fatal form of leishmaniasis which affects 70 countries, worldwide. Increasing drug resistance, HIV co-infection, and poor health system require operative vaccination strategy to control the VL transmission dynamics. Therefore, a holistic approach is needed to generate T and B memory cells to mediate long-term immunity against VL infection. Consequently, immunoinformatics approach was applied to design the multi-epitope based subunit vaccine construct consisting of B and T cell epitopes. Further, the physiochemical characterization was performed to check the aliphatic index, theoretical PI, molecular weight, and thermostable nature of vaccine construct. The allergenicity and antigenicity were also predicted to ensure the safety and immunogenic behavior of final vaccine construct. Moreover, homology modeling, followed by molecular docking and molecular dynamics simulation study was also performed to evaluate the binding affinity and stability of receptor (TLR-4) and ligand (vaccine protein) complex. This study warrants the experimental validation to ensure the immunogenicity and safety profile of presented vaccine construct that may be helpful to control VL infection.
Michigan State University, USA
Antibiotic resistance is one of the biggest threats to global health, food security, and development today. Global consumption of antibiotics has increased nearly 40% in the last decade. The incredible rapid resistance of antibiotic resistance which is taking place worldwide is not only a serious threat to the practice of modern medicine, but equally important, a threat to global public health. This urgent issue is so alarming that it caught the attention of G-20 Summit in both China (2016) and Germany (2017), let alone the U.N. Assembly in 2016 had called for a special meeting of “superbugs” which focused on the escalating drug resistance with respect to the sexually transmitted disease gonorrhea and carbapenem resistant Enterobacteriaceae. While the causes of antibiotic resistance are complex, certainly human behavior play a significant role in the spread of antibiotic resistant genes. In addition to the human behavior, the drivers of resistance include agriculture sector, animal husbandry, household and industry–these factors contribute significantly to the spread of the resistant genes within the ecosystem. Such resistant mechanisms are continuously emerging globally, which threatens our ability to treat common infections, resulting in increased death, disability and costs. Since the development and clinical use of penicillins, nearly 1000 resistant-related beta-lactamases that inactivate various types of antibiotics have been identified. There is also a global concern about the emergence of antibiotic resistant carried by the healthy individuals, the commensal bacteria. The CDC and WHO surveillance data shows that the resistance in E. coli is generally and consistently the highest for antibacterial agents in both human and veterinary medicine. Within communities, resistant bacteria circulate from person to person or from animals and environment to person, or vice versa. With 1 billion people travelling each year, bacteria is becoming more mobile. The bacterial resistance can kill 700,000 worldwide each year and itʼs been estimated to kill 10 million by 2050. The WHO estimates 78 million people a year get gonorrhea, an STD that can infect the genitals, rectum and throat - there is a widespread resistance to the first-line medicine ciprofloxacin as well as increasing resistance to azithromycin. The emergence of resistance to last-resort treatments known as extended-spectrum cephalosporins (ESCs) is now eminent. The five riskiest superbugs are recognized as (1) the original one: Staphylococcus Aureus (MRSA), (2) the hospital lurkers: Clostridium Difficile and Acinetobacter, (3) the food borne pathogens: Escherichia Coli and Salmonella, (4) The sexually-transmitted infections: Gonorrhea and Chlamydia, and (5) TB. India is a typical example of encountering the deadly bacterial resistance. The discovery of the New Delhi metallo-beta-lactamase-1 (NDM-1) which disables almost all antibiotics directed against it was turning point in the rapid emergence of blaNDM-1 gene which was first identified in 2008 in people who had traveled in India or sought medical care in South Asia. The gene for NDM-1 travels on a plasmid, an extra-chromosomal loop of DNA that can be traded freely among bacteria. So far, it has been found a variety of bacterial species that carry NDM-1 particularly in the gut bacteria, can cause serious infections in vulnerable hospital patients in India, South Asia, South Africa and the UK. There are two major routes of spread for the bacteria; hospital and the community. In hospital infections, bacteria carrying NDM-1 move from person to person when patients who have received many antibiotics, develop diarrhea and traces of feces contaminate surfaces, equipment and healthcare workersʼ hands. In community infections, the bacteria carrying the enzyme pass from person to person when traces of feces contaminate municipal water supplies–and with a large percentage of the population lacking any access to sanitation. Public Health Foundation of India believes that 60,000 infants per year are dying of drug-resistant infections due to NDM-1. In addition, tourists can pick up antibioticresistance genes in just 2-3 days. Currently, India is facing with two antibiotic resistant genes what carry NDM-1; E. coli and Klebsiella. The discovery mrc-1 gene in China which is being transferred between Klesbsiella pneumoniae and E. coli further compounded the global burden of antibiotic resistance, which has already spread to the neighboring countries. In the animal husbandry and agricultural sectors of China, the demand for the antibiotics to reach almost 12,000 tons per year. The high prevalence of the mrc-1 gene in E. Colisamples both in animals and raw meat, with the number of positive-testing samples are increasing each year in China. On average, more than 20 percent of bacteria in the animal samples and 15 percent of the raw meat samples carried the mrc-1 gene. Numerous European countries have reported the existence of mrc-1 gene in the isolates from human, isolates from animals used for food, isolates from food, and isolated from the environment.
In conclusion, antibiotic resistance can affect anyone, of any age, in any country primarily due to misuse of antibiotics in humans and animals is accelerating the process. A growing number of infections–such as pneumonia, tuberculosis, gonorrhea, and salmonellosis–are becoming harder to treat as the antibiotics used to treat them become less effective which may leads to longer hospital stays, higher medical costs and increased mortality. There is an urgent need between research universities and industry aimed at developing novel antimicrobial agents to save the practice of modern medicine, in particular, infectious diseases.
Hawler Medical University, Iraq
Background and objective: Pseudomonas aeruginosa is frequently responsible for the outbreaks of hospital-acquired infection. Multidrug resistance (MDR) in P. aeruginosa has appeared as an issue of great concern with the emergence of Metallo-β-lactamase (MBL)-producing isolates, which showed to be the primary resistance mechanism in carbapenem-resistant P. aeruginosa. Strains producing these enzymes have increased in frequency over the past few years and have been responsible for prolonged nosocomial outbreaks. The aim of the present study was detection of MBL-producing P. aeruginosa isolates from patients and then evaluate antibiotic guide for clinicians in recommending accurate antibiotic and further controlling hospital infection.
Methods: A total of 98 consecutive isolates of P. aeruginosa causing infections were isolated from various clinical samples. We applied Vitek-2 automated system as a panel of antimicrobial agents. Imipenem-EDTA combined disk test (CDT) and modified Hodge test (MHT) for phenotypic detection of MBL-producing isolates were recommended; it was performed on all P.aeruginosa isolates showing resistance to imipenem.
Results: Out of 98 isolates of P.aeruginosa screened for imipenem (IMP) susceptibility, only 72 isolates (73%) showed IMPresistance and the remaining 26 (26%) were sensitive to imipenem. Around 63 (87.5%) of the IMP-resistant isolates were able to produce MBLs as determined by CDT, while 30 (41.6) showed positive results in MHT. The resistance to antibiotics tested was significantly higher (P <0.001) with MBL producing P. aeruginosa isolates compared to non-MBL producers.
Conclusions: Our results support the belief that MBL producing-P.aeruginosa is being discovered in our region at an alarming rate. The prevalence of MDR to the antibiotics among MBL-producing P. aeruginosa isolates was established. CDT is a reliable screening test for detection of MBL. Colistins are recommended for the treatment of severe infections caused by these organisms.
Keywords: P. aeruginosa, MBL, Multi-drug resistance, phenotypic detection
1University of the Punjab, Pakistan
2National Textile University, Pakistan
In this work toxic Selenite which is also highly soluble is transformed to Selenium a less toxic element Selenium through biotic transformation using different strains of bacterial like Pseudomonas, Exiguobacterium sp, Bacillus Subtilis and licheniformis. Selenium can exists in different forms like reduced form (Selenide, Se2-), water dissolved form (Selenite, So3-2/Selenate, SeO4-2) and in the form of element (Seo). Different physical parameters were changed for optimizing conditions like different concentrations of Selenium (Se) varying from 200 to 400 and finally to 600 µg ml-1), temperature, pH, aeration along with incubation time for high reduction of Selenite. It was found that Selenite reduction rate was increased by increasing pH. It was found that at pH 3 around 15-33% Selenite was reduced and this trend kept on increasing to 28-90% at pH 9. For evaluating optimum temperature for Selenite reduction three levels of temperature were selected (32 °C, 37 °C and 42 °C) were selected. The Selenite reduction was found at different temperatures and the results showed that for optimum reduction of Selenite all strains posses varying preferences. The reduction in Selenite was also checked at different concentrations of Selenite and it was found that maximum reduction of Selenite was observed at lower concentration. This study concluded that in aerobic and anaerobic environment Se can be remediated by using selenite reducing bacteria.
Biography:
Saima Javed is doing Ph.D from University of the Punjab (Department of Microbiology & Molecular genetics). Currently she is working on screening of biosurfactants and their role in oil biodegradation. She has worked in diagnosis of dengue serotypes by molecular techniques and prevalence of Dengue in Pakistan. She has also worked on heavy metals biotransformation, bioremediation, biodegradation, and phytoremediation. In future she is interested to join the group in Pakistan to work for the development of vaccination of Dengue virus.
Hopital Edouard Herriot, France
Guidelines for the prevention of surgical site infection issued by the French Society of Hygiene Hospital1 and World Health Organization2 are recently in favor of a use of the chlorhexidine hydroalcoholic (CHX-OH) solution. The objective of the study was to develop new colored CHX-OH solution enabling skin staining mandatory for efficient pre-operative antisepsis. The stability of CHX-OH solution supplemented with various dyes (sunset yellow, eosin, erythrosin) was examined immediately after mixing ingredients in order to detect the absence of sedimentation, flocculation or discoloration. The most stable colored CHX-OH solution was stored at 25 °C ± 2 °C, 60% ± 5% RH and 40 °C ± 2 °C, 75% ± 5% RH for 24 months. Pharmaceutical control, in accordance with Good Preparation Practices and European Pharmacopoeia, included the organoleptic characteristics, the dosage of the active ingredients and a microbiological control. The determinations of In-vitro antiseptic efficacy according to AFNOR standards (NF EN 1040, NF EN 1275), and the efficacy of the In-vivo coloring properties on white and black skin by using a colorimetric skin probe have completed this study. The tests demonstrated a better stability of CHX-OH (0.5 to 2.5% (w/v)) and eosin (0.02 to 0.05% (w/v)) solutions. All controls were satisfactory over 24 months at 25 °C and up to 6 months at 40 °C, but due to a random potential instability, the expiry date was fixed preferentially at 18 months. The antiseptic activity of this solution complied with AFNOR standards. Colorimetric measurements have shown that eosin has the most intense staining property, visible on white and black skin. CHX-OH and eosin solution, having many clinical and technical advantages, has been upgraded by an international patent WO2017021667 (A1)3.
References:
1 Societe française dʼhygiene hospitalière. Antisepsie de la peau saine avant un geste invasif chez lʼadulte Recommandations pour la pratique Clinique, 2016
2 World Health Organization. Global guidelines for the prevention of surgical site infection, 2016.
3 Hydroalcoholic solutions coloured with chlorhexidine, methods for the production thereof, and uses of same. S. Filali, F. Pirot, L. Tall, C. Pivot. Depôt de brevet WO2017021667 (A1), FR3039768 (A1), 2015-08-06
Biography:
Samira Filali is a hospital pharmacist specializing in the preparation and control of the drug. His research in the pharmaceutical galenical research and development laboratory focuses on the development of new techniques for the administration of active ingredients.
University of Benin, Nigeria
Neonatal sepsis remains a major cause of morbidity and mortality in neonates. The prevalence of bacterial isolates in neonates admitted due to sepsis together with mothers and their susceptibility to routinely used antibacterial agents were investigated. Ethical Approval was obtained from the Hospital Management Board while informed consents were obtained from their parents. Forty-five (45) saliva samples from neonates (with signs and symptoms of sepsis admitted at the neonatal unit of Central Hospital, Benin City, Nigeria) were obtained. Vaginal swab samples were also obtained from their mothers to attain a total of ninety samples i.e forty-five neonate-mother saliva-vaginal swab sample pairs. Samples were immediately transported to the Laboratory and processed using standard microbiological protocols. All samples showed significant bacterial growth. At least one similar bacteria were recovered from each neonate-mother pair. Bacteria isolated include Staphylococcus aureus, Klebsiellaoxytoca, Klebsiellaspp., Escherichia coli, Enterobacter spp., Proteus miriabilis. Acinetobacter sp.,Proteus vulgaris, Pseudomonas aeruginosa, Streptococcus agalaticeae, Citrobacter sp. and Streptococcus pneumoniae. Both neonatal and maternal isolates were sensitive to unacin, azithromycin, cefotaxime and cefuroxime. Bacterial isolates also showed varying degrees of resistance to bactericidal action of normal serum. Isolates also produced haemolysin. This study gives important insight to the role of saliva in bacteriological analysis of sepsis and has implications for neonatal survival.
Keywords: Bacterial, neonatal, antibiotics.
Biography:
Samuel Abumhere Aziegbemhin is a Lecturer in the Department of Microbiology, University of Benin, Nigeria. He obtained M.Sc. degrees in Medical Microbiology from the University of Benin in 2014. His research focus is in Neonatal infections, Medical Microbiology and infectious diseases. Mr. Aziegbemhin has authored four journal publications with seasoned researchers. He has done some studies on bacterial infections in neonates. Aziegbemhin is a winner of the Brenda Howe Africa Scholarship 2012 Nottingham Trent University, United Kingdom. He is also a recipient of the Early Career Academic Grant of The Association of Commonwealth Universities (ACU) 2016.
Shivaji University, India
We have reported ecofriendly extracellular and cost- effective synthesis of Silver Nanoparticles by isolated Bacillus marisflavi YCISMN5 for the first time. The AgNps were charachterized by UV-Vis Spectroscopy, XRD, FTIR FE-SEM EDS, and DLS. The UV- Visible spectroscopic analysis showed strong surface Plasmon resonance(SPR) peak at 434 nm. XRD analysis revealed the crystalline nature of AgNps with Fcc lattice. The FTIR analysis revealed the role of biomolecules in the formation of AgNps. FESEM revealed the size of AgNps in the range of 20-30 nms. EDS confirmed the formation of metallic AgNps. Average particle size calculated using DLS measurements was found to be approximately 27 nm.
Hospital acquired infections are a major problem, doctors and patients have to face in day to day life. AgNps have received more importance than metallic silver due to their increased antimicrobial activity at nanoscale.
Representative pathogens causing nosocomial infections, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923, and Escherichia coli ATCC 25922, were treated with the biosynthesized AgNps to assess their antibacterial activity, singly and in combination with third generation antibiotics.
Results: It was observed that AgNps showed good antibacterial activity against Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli than standard antibiotic, whereas enhanced antibacterial activity of AgNps in combination with cefperazone was observed against Pseudomonas aeruginosa as compared to standard antibiotic and the zone of inhibition was found to be increased by 0.13 fold(P≤.05). This result seems to be significant in designing strategies for curing Pseudomonas infection.
Biography:
Shivangi Shivraj Kanase is working as an Associate Professor in P.G. Department of Microbiology. She has a teaching experience of twenty- seven years. She completed her Ph.D. from Shivaji University in 2007under FIP. As a research guide she is guiding six students for their Ph.D. Her areas of research include Nanobiotechnology, Endophytes from medicinal plants and identification of their antifungal and antibacterial compounds, and Bioremidiation-Textile Dye degradation. Published one National and five International papers. Worked on two Minor research projects sanctioned by U.G.C. She has participated in approximately thirty seven National and International Conferences and three hands on training workshops. She and her student have won gold medal for best paper presentations at international conferences.
University of Munster, Germany
In the last decades the increasing rate of multidrug-resistance to classical antibiotics has driven research towards identification of other means to fight bacterial infections. Increasing evidence infers that bacteria being internalized into epithelial cells are responsible for a high incidence of recurrent infections.
Our previous studies showed that cell-penetrating peptides (CPP) could mediate the internalization of antimicrobial compounds into the cytoplasm of host cells followed by the efficient killing of intracellular pathogens. Here, we investigated whether virulence factors, such as bacteria-derived peptidoglycan-degrading proteins, fused to a virus-derived CPP can be used against intracellular pathogenic bacteria generating novel potential therapeutic tools to treat recurrent infectious diseases. Furthermore, these antimicrobial compounds are not inhibited by traditional antibiotic resistance mechanisms.
Fusion proteins of recombinant variants of selected peptidoglycanases with CPP have been generated. The antimicrobial activity of these recombinant proteins was assessed by several bioassays such as turbidity assay, membrane impairment assay, spot assay, and MIC assay against various Gram-positive pathogens including methicillin resistant S. aureus (MRSA) and also Gram-negative pathogens.
Our results confirm and underline the potential of CPP to deliver antimicrobial agents. However, peptidoglycan-degrading enzymes turned out to be species specific and when over expressed can be quite toxic to the host bacteria. Taken together, fusions of CPP with peptidoglycan-degrading proteins or homologues to lysozyme, which maintain the high enzymatic activity, may represent novel, innovative antimicrobials.
Keywords: Antimicrobial compounds; cell-penetrating peptides; peptidoglycan-degrading proteins, intracellular pathogens.
CSIR–National Environmental Engineering Research Institute, India
This study aims to explore the microbial diversity of ruminant organisms Bostaurus (cow) and Bubalus bubalis (buffalo) under different feeding scenario and screen well-adapted rumen microbiome to select and characterize potentially enhanced lignocellulolytic enzyme-producing microbes. The microbiota of green roughage fed cow (NDC_GR), buffalo (NDB_GR) and dry roughage fed cow (NDC_DR), buffalo (NDB_DR) were explored using high-throughput sequencing. Analysis revealed the presence of highly complex bacterial community, with the maximal depiction by the phyla Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria. Moreover, bacteria were isolated and screened for exo-1, 4-β-glucanase, β-glucosidase, α-glucuronidase, endo-1, 4-β-xylanases, arabinosidase and α-galactosidase activities using differential chromogenic substrates. Forty- five isolates that showed multienzymatic activities were identified using 16S rDNA sequencing. Genome annotation of P. polymyxa ND24, capable of hydrolyzing various plant biomasses further revealed efficient lignocellulolytic machinery comprising ten cellulases and twenty-one hemicellulases, implicated for sustainable biomass hydrolysis. Present approach establishes the rumen microbiome as a substantial reservoir of industrially important bacterial strains for upgrading the possibility of lignocellulose utilization for the ‘environmental friendly’ approach of second-generation biofuels.
Biography:
Varsha Bohra is a Ph.D. scholar at CSIR–National Environmental Engineering Research Institute, Nagpur, India. She received her masterʼs degree in Microbiology from Pt. Ravisankar Shukla University, Raipur. She has been awarded INSPIRE fellowship from department of science and technology, India. Her current research focuses on “the metagenomic study of rumen microbiome for exploring lignocellulolytic enzymes”. She has authored 2 papers in peerreviewed scientific journals (Applied Biochemistry and biotechnology, 3 Biotech) and attended three international conferences (BRSI, India; AMI, India).